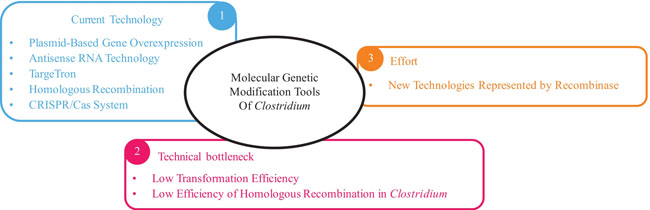
Clostridium are Gram-positive, strictly anaerobic, endospore-forming bacteria that produce a variety of chemicals, including butanol, which is now a promising new biofuel. Improving the fermentation titer and yield of Clostridium by genetic modification has always been an important challenge that needs to be broken through, but it has long been hindered by the limitation of genetic manipulation tools of Clostridium. In recent years, with the continuous development of molecular biology, gene editing tools for Clostridium have been continuously developed. Many genetic manipulation tools such as plasmid-based gene overexpression, antisense RNA technology, transposon-based mutagenesis, group Ⅱ intron-mediated gene inactivation, and homologous recombination-based or CRISPR/Cas-mediated gene editing technology have been developed. Various operations such as target gene insertion, deletion, substitution, point mutation, and gene expression level regulation have been accomplished in Clostridium. In this review, we summarize the research progress in the molecular genetic modification tools of Clostridium, and especially discuss the potential application of new technologies, such as recombinase-based gene editing technology. Although the application of the recombinase system in Clostridium is rarely reported and discussed, the future application value and significance of this technology should be paid attention to. In the future, optimization of the existing molecular genetic modification technologies in Clostridium is still imperative, such as overcoming the low efficiency of homogeneous recombination in Clostridium, improving the stability and transformation efficiency of plasmids, solving the off-target problem of antisense RNA technology and type Ⅱ intron technology, reducing the toxicity of Cas9 protein, and so on. At the same time, new gene editing technologies should be developed, focusing on emerging technologies including CRISPR/Cas-mediated multi-locus editing systems, phage recombinase-mediated multiplex genome editing, targeted or random multi-copy gene integration, and so on. It is believed that with the development and improvement of genetic modification tools, Clostridium will be able to fully each its potential biorefinery capacity and make an important contribution to the green biosynthesis of bioenergy and bio-based chemicals.