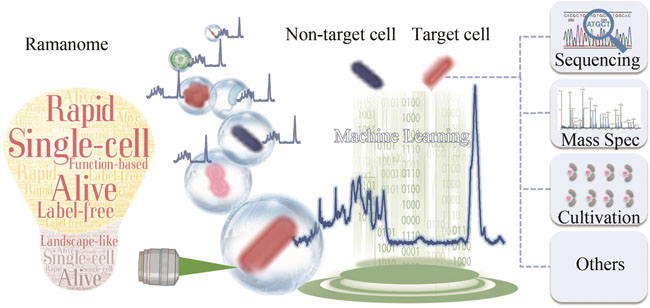
In synthetic biology, methodological innovations in sequencing, editing and synthesis of genes and whole genomes have resulted in unprecedented development in "design and manufacturing of genotypes". On the other hand, "testing of cellular phenotypes and functions" has increasingly become one of major bottlenecks. Single-cell technologies have tremendous impacts and potentials in rapid testing of cellular phenotypes and functions. However, such single-cell methods should allow non-invasive live-cell probing, be label-free, provide landscape-like phenotype sorting, distinguish complex functions, operate with high speed, sufficient throughput and low-cost, and finally, be able to integrate with downstream omics analysis. Raman spectroscopy has all the above features, and can provide information on the chemical composition and molecular structure of single cells, making it an efficient single-cell phenotyping technology. In this review, we first introduce the concept of Ramanome and Ramanome-based phenotyping technologies, including detecting and quantifying products, measuring profiles of substrates and metabolites, discriminating cell types or states, and characterizing stress response and modeling environmental changes. We then summarize the development of existing Raman-activated cell sorting (RACS) platforms in phenotyping and sorting of cell factories such as including spontaneous Raman, resonance Raman, and coherent Raman, the modes for acquiring Raman signals including static modes on dry slice and in liquid as well, flow modes by trap-free and trap-and-release manners, and principles for target cells sorting including Ejection by pulsed laser, dragging by optical tweezer, and sorting by microfluidics operation and droplets. We also highlight the applications of different RACS platforms, including the sorting of carotenoid-producing yeast and cyanobacteria cells, astaxanthin (AXT)-hyperproducing microalgae cells, triacylglycerol (TAG)-producing yeast cells, etc. Finally, challenges with single cell Raman spectroscopy (SCRS) in the phenotyping and sorting of synthetic cells and their perspectives are outlined and discussed. We propose that SCRS will bridge phenotypes and genotypes in science and technologies through coupling with downstream high-throughput cell sorting and omics profiling. This bridge will lead to novel and creative solutions to high-throughput, landscape-like testing and screening of synthetic cells. Moreover, it will fulfill the promise of Raman spectroscopy-enabled single-cell "phenome-genome" as a new type of biological big-data, and accelerate the pace of "data-driven" synthetic biology.