Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (1): 30-46.DOI: 10.12211/2096-8280.2022-055
• Invited Review • Previous Articles Next Articles
Recent progress in computational tools for designing editing sequences used in microbial genetic manipulations
YANG Yi1, MAO Yufeng1, YANG Chunhe1,2, WANG Meng1, LIAO Xiaoping1, MA Hongwu1
- 1.Key Laboratory of Systems Microbial Biotechnology,Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,Tianjin 300308,China
2.College of Biotechnology,Tianjin University of Science & Technology,Tianjin 300457,China
-
Received:2022-10-03Revised:2022-11-23Online:2023-03-07Published:2023-02-28 -
Contact:MA Hongwu
面向微生物遗传操作的编辑序列设计工具的研究进展
杨毅1, 毛雨丰1, 杨春贺1,2, 王猛1, 廖小平1, 马红武1
- 1.中国科学院天津工业生物技术研究所中国科学院系统微生物工程重点实验室,天津 300308
2.天津科技大学生物工程学院,天津 300457
-
通讯作者:马红武 -
作者简介:杨毅(1986—),男,博士研究生。研究方向为生物信息学、合成生物学、代谢工程。杨毅 (1986—),男,博士研究生。研究方向为生物信息学、合成生物学、代谢工程。E-mail:yangyi@tib.cas.cn马红武 (1970—),男,博士,研究员,博士生导师。研究方向为计算生物学、系统生物学等。E-mail:ma_hw@tib.cas.cn -
基金资助:国家重点研发计划“合成生物学”重点专项(2018YFA0902900);国家自然科学基金(32101186);中国科学院青年创新促进会;天津市合成生物技术创新能力提升行动项目(TSBICIP-PTJS-001)
CLC Number:
Cite this article
YANG Yi, MAO Yufeng, YANG Chunhe, WANG Meng, LIAO Xiaoping, MA Hongwu. Recent progress in computational tools for designing editing sequences used in microbial genetic manipulations[J]. Synthetic Biology Journal, 2023, 4(1): 30-46.
杨毅, 毛雨丰, 杨春贺, 王猛, 廖小平, 马红武. 面向微生物遗传操作的编辑序列设计工具的研究进展[J]. 合成生物学, 2023, 4(1): 30-46.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-055
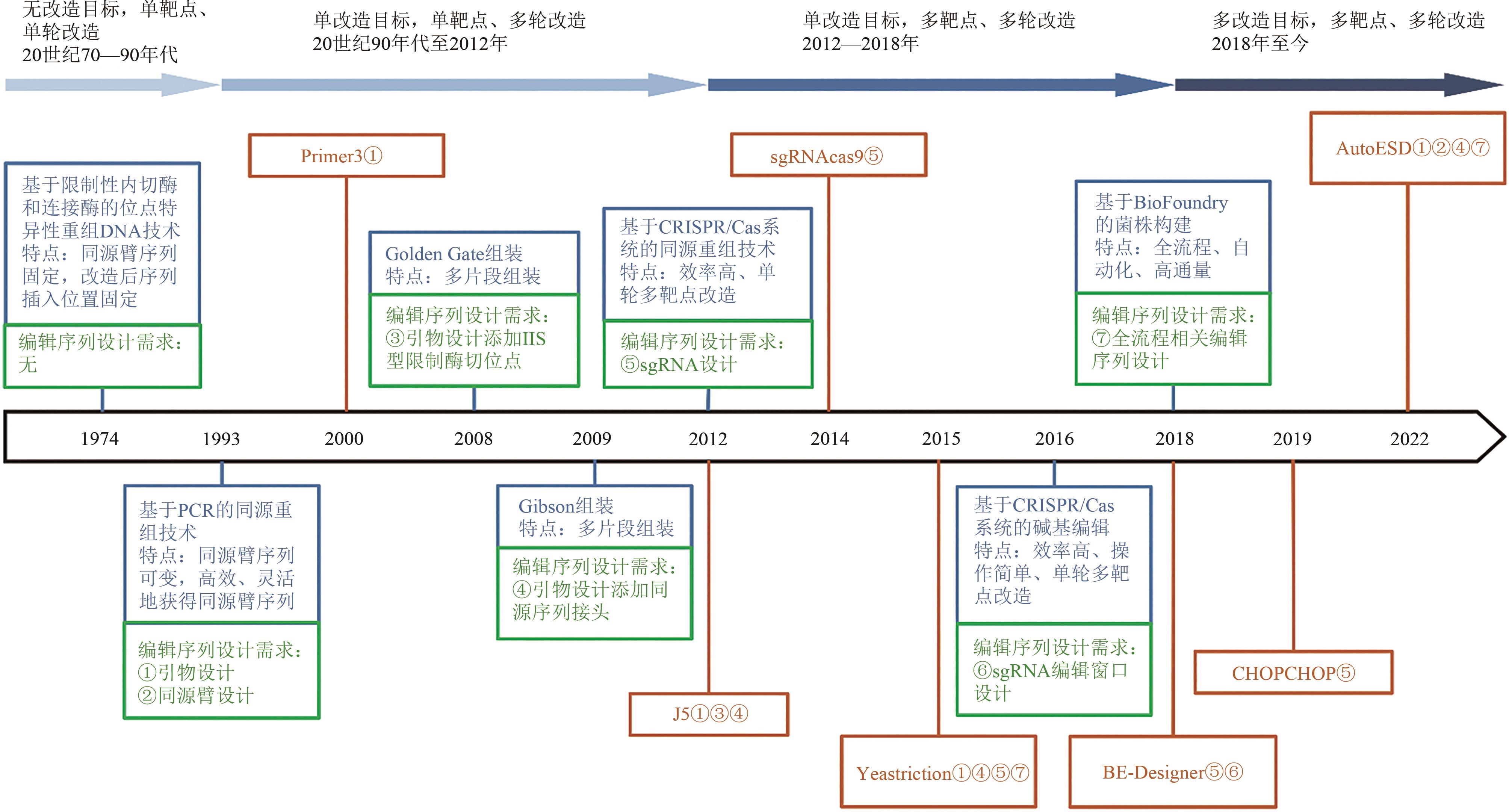
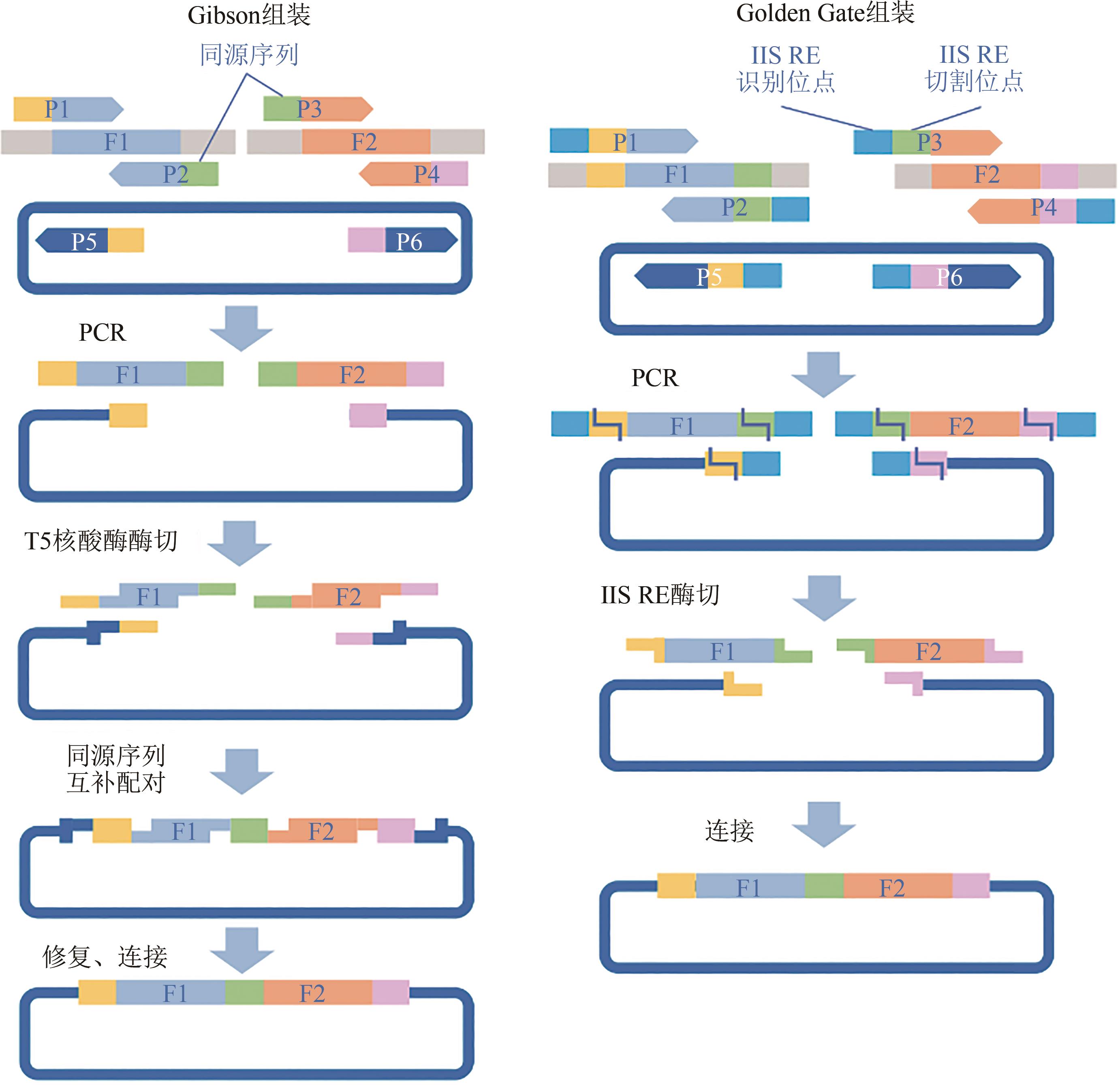
Fig. 1 Development of genetic manipulation technologies and corresponding editing sequences and tools for editing sequence design(Blue rectangle: Genetic manipulation technologies; Green rectangle: Editing sequence design requirements; Brown rectangle: Editing sequence design tools; Brown serial numbers indicates that the editing sequence design tools can address the corresponding editing sequence design requirements)
| 工具 | 基于Primer3 | 特异性检验 | 批量设计 | 功能特色 | 可用性 | URL |
|---|---|---|---|---|---|---|
| Primer3[ | — | 否 | 否 | 基础引物设计 | 在线/图形界面、离线/命令行 | https://bioinfo.ut.ee/primer3/ |
| BatchPrimer3[ | 是 | 否 | 是 | 批量引物设计 | 在线/图形界面 | http://probes.pw.usda.gov/cgi-bin/batchprimer3/batchprimer3.cgi |
| ConservedPrimer2.0[ | 是 | 是 | 是 | Sanger测序引物设计 | 在线/图形界面 | https://probes.pw.usda.gov/ConservedPrimers/index.html |
| PrimerDesign-M[ | 否 | 是 | 是 | Sanger测序引物设计 | 在线/图形界面 | www.hiv.lanl.gov/content/sequence/PRIMER_DESIGN/primer_design.html |
| Primer-BLAST[ | 是 | 是 | 否 | RT-PCR引物设计 | 在线/图形界面 | https://www.ncbi.nlm.nih.gov/tools/primer-blast/ |
| PCR designer[ | 否 | 否 | 否 | SNP分析引物设计 | 在线/图形界面 | http://primer1.soton.ac.uk/primer.html |
| PRIMEGENS-v2[ | 是 | 否 | 是 | 剪接变体引物设计 | 在线/图形界面、离线/命令行 | http://primegens.org/ |
| PrimerSeq[ | 是 | 否 | 是 | 剪接变体引物设计 | 离线/图形界面 | https://primerseq.sourceforge.net/ |
| MSP-HTPrimer[ | 是 | 否 | 是 | DNA甲基化分析引物设计 | 离线/命令行 | https://sourceforge.net/projects/msp-htprimer/ |
| MPD[ | 否 | 是 | 是 | 多重PCR引物设计 | 离线/命令行 | https://wingolab-org.github.io/mpd-c/ |
| Mutation Maker[ | 是 | 是 | 是 | 多重PCR引物设计、氨基酸突变引物设计 | 离线/命令行 | https://github.com/ra100/Mutation_Maker |
| PerlPrimer[ | 否 | 否 | 是 | Sanger测序引物设计、RT-PCR引物设计、开放阅读框(ORF)搜索及相关引物设计 | 离线/图形界面 | https://perlprimer.sourceforge.net/ |
Table 1 Major primer design tools with different functions
| 工具 | 基于Primer3 | 特异性检验 | 批量设计 | 功能特色 | 可用性 | URL |
|---|---|---|---|---|---|---|
| Primer3[ | — | 否 | 否 | 基础引物设计 | 在线/图形界面、离线/命令行 | https://bioinfo.ut.ee/primer3/ |
| BatchPrimer3[ | 是 | 否 | 是 | 批量引物设计 | 在线/图形界面 | http://probes.pw.usda.gov/cgi-bin/batchprimer3/batchprimer3.cgi |
| ConservedPrimer2.0[ | 是 | 是 | 是 | Sanger测序引物设计 | 在线/图形界面 | https://probes.pw.usda.gov/ConservedPrimers/index.html |
| PrimerDesign-M[ | 否 | 是 | 是 | Sanger测序引物设计 | 在线/图形界面 | www.hiv.lanl.gov/content/sequence/PRIMER_DESIGN/primer_design.html |
| Primer-BLAST[ | 是 | 是 | 否 | RT-PCR引物设计 | 在线/图形界面 | https://www.ncbi.nlm.nih.gov/tools/primer-blast/ |
| PCR designer[ | 否 | 否 | 否 | SNP分析引物设计 | 在线/图形界面 | http://primer1.soton.ac.uk/primer.html |
| PRIMEGENS-v2[ | 是 | 否 | 是 | 剪接变体引物设计 | 在线/图形界面、离线/命令行 | http://primegens.org/ |
| PrimerSeq[ | 是 | 否 | 是 | 剪接变体引物设计 | 离线/图形界面 | https://primerseq.sourceforge.net/ |
| MSP-HTPrimer[ | 是 | 否 | 是 | DNA甲基化分析引物设计 | 离线/命令行 | https://sourceforge.net/projects/msp-htprimer/ |
| MPD[ | 否 | 是 | 是 | 多重PCR引物设计 | 离线/命令行 | https://wingolab-org.github.io/mpd-c/ |
| Mutation Maker[ | 是 | 是 | 是 | 多重PCR引物设计、氨基酸突变引物设计 | 离线/命令行 | https://github.com/ra100/Mutation_Maker |
| PerlPrimer[ | 否 | 否 | 是 | Sanger测序引物设计、RT-PCR引物设计、开放阅读框(ORF)搜索及相关引物设计 | 离线/图形界面 | https://perlprimer.sourceforge.net/ |
| 工具 | 基于Primer3 | 支持的DNA片段组装技术 | 批量设计 | 可用性 | URL |
|---|---|---|---|---|---|
| J5[ | 是 | Golden Gate/Gibson/ CPEC/SLIC | 是 | 在线/图形界面 | https://j5.jbei.org/ |
| Geneious | 否 | Gibson | 是 | 在线/图形界面 | https://www.geneious.com/ |
| Raven[ | 否 | Gibson | 是 | 在线/图形界面 | http://ravencad.org/ |
| MCDS[ | 否 | GoldenGate/Gibson/ SLIC/Gateway | 否 | 离线/命令行 | https://github.com/errisy/MCDS |
| NEBbridge Golden Gate[ | 未知 | Golden Gate | 是 | 在线/图形界面 | https://goldengate.neb.com/ |
| iBioCAD GGA[ | 未知 | Golden Gate | 是 | 在线/图形界面 | https://ibiocad.igb.illinois.edu/ |
| PlasmidMaker[ | 是 | Golden Gate/Gibson | 是 | 在线/图形界面 | https://biofoundry.web.illinois.edu/ |
Table 2 Representative design tools for DNA assembly
| 工具 | 基于Primer3 | 支持的DNA片段组装技术 | 批量设计 | 可用性 | URL |
|---|---|---|---|---|---|
| J5[ | 是 | Golden Gate/Gibson/ CPEC/SLIC | 是 | 在线/图形界面 | https://j5.jbei.org/ |
| Geneious | 否 | Gibson | 是 | 在线/图形界面 | https://www.geneious.com/ |
| Raven[ | 否 | Gibson | 是 | 在线/图形界面 | http://ravencad.org/ |
| MCDS[ | 否 | GoldenGate/Gibson/ SLIC/Gateway | 否 | 离线/命令行 | https://github.com/errisy/MCDS |
| NEBbridge Golden Gate[ | 未知 | Golden Gate | 是 | 在线/图形界面 | https://goldengate.neb.com/ |
| iBioCAD GGA[ | 未知 | Golden Gate | 是 | 在线/图形界面 | https://ibiocad.igb.illinois.edu/ |
| PlasmidMaker[ | 是 | Golden Gate/Gibson | 是 | 在线/图形界面 | https://biofoundry.web.illinois.edu/ |
| 工具 | 编辑效率预测方法 | 特异性预测方法 | PAM | 支持的改造类型 | URL |
|---|---|---|---|---|---|
| E-CRISP[ | 假设先验 | 假设先验 | NGG | 敲除、激活、抑制 | http://www.e-crisp.org/E-CRISP/index.html |
| CRISPR-ERA[ | 假设先验 | 假设先验 | NGG | 敲除、激活、抑制 | http://CRISPR-ERA.stanford.edu |
| EuPaGDT[ | 假设先验 | 假设先验 | NGG、TTTN、NGA等 | 敲除 | http://grna.ctegd.uga.edu/ |
| CRISPOR[ | 假设先验/机器学习 | 假设先验 | NGG、TTTN、NGA等 | 敲除 | http://crispor.tefor.net/ |
| CHOPCHOP[ | 假设先验/机器学习 | 假设先验 | NGG、TTTN、NGA等 | 敲除、敲入、激活、抑制 | http://chopchop.cbu.uib.no/ |
| CRISPETa[ | 假设先验 | 数据库搜索 | NGG | 敲除 | http://crispeta.crg.eu |
| CRISPR-P[ | 假设先验 | 假设先验 | NGG、NRG、TTN等 | 敲除 | http://crispr.hzau.edu.cn/CRISPR2/ |
| CRISPRscan[ | 机器学习 | 假设先验 | NGG、TTTN、NGN等 | 敲除 | https://www.crisprscan.org |
| WU-CRISPR[ | 机器学习 | 假设先验 | NGG | 敲除 | http://crisprdb.org/wu-crispr/ |
| CRISTA[ | 机器学习 | 机器学习 | NGG | 敲除 | https://crista.tau.ac.il/ |
| Elevation[ | 机器学习 | 机器学习 | NGG | 敲除 | https://crispr.ml/ |
Table 3 Representative hypothesis-driven and learning-based sgRNA design tools
| 工具 | 编辑效率预测方法 | 特异性预测方法 | PAM | 支持的改造类型 | URL |
|---|---|---|---|---|---|
| E-CRISP[ | 假设先验 | 假设先验 | NGG | 敲除、激活、抑制 | http://www.e-crisp.org/E-CRISP/index.html |
| CRISPR-ERA[ | 假设先验 | 假设先验 | NGG | 敲除、激活、抑制 | http://CRISPR-ERA.stanford.edu |
| EuPaGDT[ | 假设先验 | 假设先验 | NGG、TTTN、NGA等 | 敲除 | http://grna.ctegd.uga.edu/ |
| CRISPOR[ | 假设先验/机器学习 | 假设先验 | NGG、TTTN、NGA等 | 敲除 | http://crispor.tefor.net/ |
| CHOPCHOP[ | 假设先验/机器学习 | 假设先验 | NGG、TTTN、NGA等 | 敲除、敲入、激活、抑制 | http://chopchop.cbu.uib.no/ |
| CRISPETa[ | 假设先验 | 数据库搜索 | NGG | 敲除 | http://crispeta.crg.eu |
| CRISPR-P[ | 假设先验 | 假设先验 | NGG、NRG、TTN等 | 敲除 | http://crispr.hzau.edu.cn/CRISPR2/ |
| CRISPRscan[ | 机器学习 | 假设先验 | NGG、TTTN、NGN等 | 敲除 | https://www.crisprscan.org |
| WU-CRISPR[ | 机器学习 | 假设先验 | NGG | 敲除 | http://crisprdb.org/wu-crispr/ |
| CRISTA[ | 机器学习 | 机器学习 | NGG | 敲除 | https://crista.tau.ac.il/ |
| Elevation[ | 机器学习 | 机器学习 | NGG | 敲除 | https://crispr.ml/ |
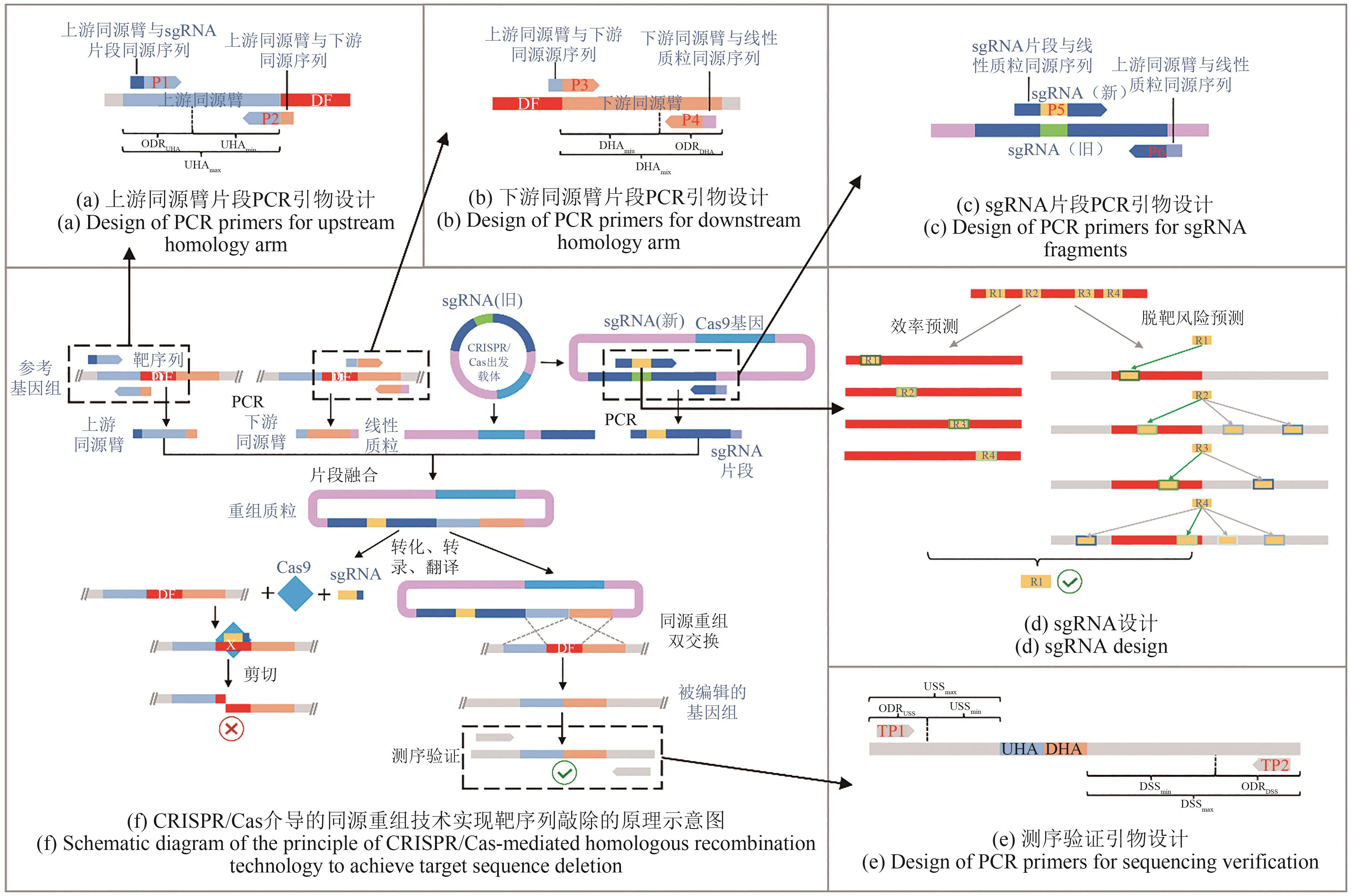
Fig. 4 Schematic diagram for CRISPR/Cas-based homologous recombination and its editing sequencesDF—Deleted fragment; P — Primer; UHAmin—Minimum length of upstream homology arm; UHAmax— Maximum length of upstream homology arm; ODRUHA—Optional design region of the upstream primer of the upstream homologous arm; DHAmin —Minimum length of downstream homology arm; DHAmax— Maximum length of downstream homology arm; ODRDHA— Optional design region of the downstream primer of downstream homologous arm; R—sgRNA (dark green and blue of the sgRNA border lines represent on-target efficiency and off-target risk, respectively); TP— Test primer; UHA— Upstream homologous arm; DHA— Downstream homologous arm; USSmin—Minimum length of upstream spacer sequence (sequence from the 3'-end of the upstream verification primer to the 5'-end of the upstream homology arm); USSmax— Maxmum length of upstream spacer sequence; ODRUSS— Optional design region of the upstream verification primer; DSSmin— Minimum length of downstream spacer sequence (sequence from the 3'-end of the downstream verification primer to the 3'-end of the downstream homology arm); DSSmax— Maxmum length of downstream spacer sequence; ODRDSS—Optional design region of the downstream verification primer
| 工具 | 编辑序列设计 | 遗传改造类型 | 是否高通量 | 可支持物种 | 可编辑位点 | web应用 |
|---|---|---|---|---|---|---|
| Yeastriction[ | 引物设计sgRNA设计 | 删除 | 否 | Saccharomyces cerevisiae(33个菌株) | 已被注释的基因 | http://yeastriction.tnw.tudelft.nl/#!/ |
| GeneTargeter[ | 引物设计sgRNA设计 | 删除、表达弱化 | 是 | Plasmodium falciparum | 已被注释的基因 | http://genetargeter.mit.edu/ |
| GEDpm-cg[ | 引物设计、同源臂设计 | 单点突变 | 是 | Corynebacterium glutamicum | 基因组任意区域 | https://gedpm-cg.biodesign.ac.cn/ |
| AutoESD[ | 引物设计、同源臂设计 | 插入、删除、替换、单点突变 | 是 | 多种原核、真核微生物 | 基因组任意区域 | https://autoesd.biodesign.ac.cn/ |
Table 4 Editing sequence design tools for the whole workflow of genome editing
| 工具 | 编辑序列设计 | 遗传改造类型 | 是否高通量 | 可支持物种 | 可编辑位点 | web应用 |
|---|---|---|---|---|---|---|
| Yeastriction[ | 引物设计sgRNA设计 | 删除 | 否 | Saccharomyces cerevisiae(33个菌株) | 已被注释的基因 | http://yeastriction.tnw.tudelft.nl/#!/ |
| GeneTargeter[ | 引物设计sgRNA设计 | 删除、表达弱化 | 是 | Plasmodium falciparum | 已被注释的基因 | http://genetargeter.mit.edu/ |
| GEDpm-cg[ | 引物设计、同源臂设计 | 单点突变 | 是 | Corynebacterium glutamicum | 基因组任意区域 | https://gedpm-cg.biodesign.ac.cn/ |
| AutoESD[ | 引物设计、同源臂设计 | 插入、删除、替换、单点突变 | 是 | 多种原核、真核微生物 | 基因组任意区域 | https://autoesd.biodesign.ac.cn/ |
| 26 | ROZEN S, SKALETSKY H. Primer3 on the WWW for general users and for biologist programmers[J]. Methods in Molecular Biology, 2000, 132: 365-386. |
| 27 | KORESSAAR T, REMM M. Enhancements and modifications of primer design program Primer3[J]. Bioinformatics, 2007, 23(10): 1289-1291. |
| 28 | YOU F M, HUO N X, GU Y Q, et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design[J]. BMC Bioinformatics, 2008, 9: 253. |
| 29 | ANDRESON R, REPPO E, KAPLINSKI L, et al. GENOMEMASKER package for designing unique genomic PCR primers[J]. BMC Bioinformatics, 2006, 7: 172. |
| 30 | BOUTROS R, STOKES N, BEKAERT M, et al. UniPrime2: a web service providing easier Universal Primer design[J]. Nucleic Acids Research, 2009, 37(): W209-W213. |
| 31 | SHEN Z Y, QU W B, WANG W, et al. MPprimer: a program for reliable multiplex PCR primer design[J]. BMC Bioinformatics, 2010, 11: 143. |
| 32 | ZIESEL A C, CHRENEK M A, WONG P W. MultiPriDe: automated batch development of quantitative real-time PCR primers[J]. Nucleic Acids Research, 2008, 36(9): 3095-3100. |
| 33 | VIJAYA SATYA R, KUMAR K, ZAVALJEVSKI N, et al. A high-throughput pipeline for the design of real-time PCR signatures[J]. BMC Bioinformatics, 2010, 11: 340. |
| 34 | ARVIDSSON S, KWASNIEWSKI M, RIAÑO-PACHÓN D M, et al. QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR[J]. BMC Bioinformatics, 2008, 9: 465. |
| 35 | YOU F M, HUO N X, GU Y Q, et al. ConservedPrimers 2.0: a high-throughput pipeline for comparative genome referenced intron-flanking PCR primer design and its application in wheat SNP discovery[J]. BMC Bioinformatics, 2009, 10: 331. |
| 36 | YOU F M, WANJUGI H, HUO N X, et al. RJPrimers: unique transposable element insertion junction discovery and PCR primer design for marker development[J]. Nucleic Acids Research, 2010, 38(): W313-W320. |
| 37 | SRIVASTAVA G P, HANUMAPPA M, KUSHWAHA G, et al. Homolog-specific PCR primer design for profiling splice variants[J]. Nucleic Acids Research, 2011, 39(10): e69. |
| 38 | TSAI M F, LIN Y J, CHENG Y C, et al. PrimerZ: streamlined primer design for promoters, exons and human SNPs[J]. Nucleic Acids Research, 2007, 35(): W63-W65. |
| 39 | PIRIYAPONGSA J, NGAMPHIW C, ASSAWAMAKIN A, et al. RExPrimer: an integrated primer designing tool increases PCR effectiveness by avoiding 3' SNP-in-primer and mis-priming from structural variation[J]. BMC Genomics, 2009, 10(): S4. |
| 40 | YOON H, LEITNER T. PrimerDesign-M: a multiple-alignment based multiple-primer design tool for walking across variable genomes[J]. Bioinformatics, 2015, 31(9): 1472-1474. |
| 41 | YE J, COULOURIS G, ZARETSKAYA I, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction[J]. BMC Bioinformatics, 2012, 13: 134. |
| 42 | KE X Y, COLLINS A, YE S. PCR designer for restriction analysis of various types of sequence mutation[J]. Bioinformatics, 2002, 18(12): 1688-1689. |
| 43 | TOKHEIM C, PARK J W, XING Y. PrimerSeq: design and visualization of RT-PCR primers for alternative splicing using RNA-seq data[J]. Genomics, Proteomics & Bioinformatics, 2014, 12(2): 105-109. |
| 44 | PANDEY R V, PULVERER W, KALLMEYER R, et al. MSP-HTPrimer: a high-throughput primer design tool to improve assay design for DNA methylation analysis in epigenetics[J]. Clinical Epigenetics, 2016, 8:101. |
| 45 | WINGO T S, KOTLAR A, CUTLER D J. MPD: multiplex primer design for next-generation targeted sequencing[J]. BMC Bioinformatics, 2017, 18(1):14. |
| 46 | HIRAGA K, MEJZLIK P, MARCISIN M, et al. Mutation maker, an open source oligo design platform for protein engineering[J]. ACS Synthetic Biology, 2021, 10(2): 357-370. |
| 47 | MARSHALL O J. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR[J]. Bioinformatics, 2004, 20(15): 2471-2472. |
| 48 | SANTALUCIA J. Physical principles and visual-OMP software for optimal PCR design[J]. Methods in Molecular Biology, 2007, 402: 3-34. |
| 49 | WANG K, LI H W, XU Y, et al. MFEprimer-3.0: Quality control for PCR primers[J]. Nucleic Acids Research, 2019, 47(W1): W610-W613. |
| 50 | KECHIN A, BOROBOVA V, BOYARSKIKH U, et al. NGS-PrimerPlex: High-throughput primer design for multiplex polymerase chain reactions[J]. PLoS Computational Biology, 2020, 16(12): e1008468. |
| 51 | RAIBAUD O, MOCK M, SCHWARTZ M. A technique for integrating any DNA fragment into the chromosome of Escherichia coli [J]. Gene, 1984, 29(1/2): 231-241. |
| 52 | KNIGHT T. Idempotent vector design for standard assembly of biobricks[R]. MIT Artificial Intelligence Laboratory, MIT Synthetic Biology Working Group, 2003. |
| 53 | ANDERSON J C, DUEBER J E, LEGUIA M, et al. BglBricks: a flexible standard for biological part assembly[J]. Frontiers in Medicine, 2010, 4(1): 1. |
| 54 | ENGLER C, KANDZIA R, MARILLONNET S. A one pot, one step, precision cloning method with high throughput capability[J]. PLoS One, 2008, 3(11): e3647. |
| 55 | HORTON R M, HUNT H D, HO S N, et al. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension[J]. Gene, 1989, 77(1): 61-68. |
| 56 | QUAN J Y, TIAN J D. Circular polymerase extension cloning of complex gene libraries and pathways[J]. PLoS One, 2009, 4(7): e6441. |
| 57 | LI M Z, ELLEDGE S J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC[J]. Nature Methods, 2007, 4(3): 251-256. |
| 58 | DE KOK S, STANTON L H, SLABY T, et al. Rapid and reliable DNA assembly via ligase cycling reaction[J]. ACS Synthetic Biology, 2014, 3(2): 97-106. |
| 59 | APPLETON E, TAO J, HADDOCK T, et al. Interactive assembly algorithms for molecular cloning[J]. Nature Methods, 2014, 11(6): 657-662. |
| 60 | SHI Z, VICKERS C E. Molecular Cloning Designer Simulator (MCDS): all-in-one molecular cloning and genetic engineering design, simulation and management software for complex synthetic biology and metabolic engineering projects[J]. Metabolic Engineering Communications, 2016, 3: 173-186. |
| 61 | PRYOR J M, POTAPOV V, KUCERA R B, et al. Enabling one-pot Golden Gate assemblies of unprecedented complexity using data-optimized assembly design[J]. PLoS One, 2020, 15(9): e0238592. |
| 62 | HAMEDIRAD M, WEISBERG S, CHAO R, et al. Highly efficient single-pot scarless golden gate assembly[J]. ACS Synthetic Biology, 2019, 8(5): 1047-1054. |
| 1 | COHEN S N, CHANG A C, BOYER H W, et al. Construction of biologically functional bacterial plasmids in vitro [J]. Proceedings of the National Academy of Sciences of the United States of America, 1973, 70(11): 3240-3244. |
| 2 | CHANG A C, COHEN S N. Genome construction between bacterial species in vitro: replication and expression of Staphylococcus plasmid genes in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 1974, 71(4): 1030-1034. |
| 3 | KLECKNER N, ROTH J, BOTSTEIN D. Genetic engineering in vivo using translocatable drug-resistance elements: new methods in bacterial genetics[J]. Journal of Molecular Biology, 1977, 116(1): 125-159. |
| 4 | GIBSON D G, YOUNG L, CHUANG R Y, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nature Methods, 2009, 6(5): 343-345. |
| 5 | ENGLER C, GRUETZNER R, KANDZIA R, et al. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes[J]. PLoS One, 2009, 4(5): e5553. |
| 6 | SCHERER S, DAVIS R W. Replacement of chromosome segments with altered DNA sequences constructed in vitro [J]. Proceedings of the National Academy of Sciences of the United States of America, 1979, 76(10): 4951-4955. |
| 7 | MURPHY K C. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli [J]. Journal of Bacteriology, 1998, 180(8): 2063-2071. |
| 8 | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| 9 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| 10 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| 11 | STORCH M, HAINES M C, BALDWIN G S. DNA-BOT: a low-cost, automated DNA assembly platform for synthetic biology[J]. Synthetic Biology, 2020, 5(1): ysaa010. |
| 12 | SI T, CHAO R, MIN Y H, et al. Automated multiplex genome-scale engineering in yeast[J]. Nature Communications, 2017, 8: 15187. |
| 13 | ENGHIAD B, XUE P, SINGH N, et al. PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction[J]. Nature Communications, 2022, 13: 2697. |
| 14 | YANG Y, MAO Y F, WANG R Y, et al. AutoESD: a web tool for automatic editing sequence design for genetic manipulation of microorganisms[J]. Nucleic Acids Research, 2022, 50(W1): W75-W82. |
| 15 | UNTERGASSER A, CUTCUTACHE I, KORESSAAR T, et al. Primer3—new capabilities and interfaces[J]. Nucleic Acids Research, 2012, 40(15): e115. |
| 16 | HILLSON N J, ROSENGARTEN R D, KEASLING J D. J5 DNA assembly design automation software[J]. ACS Synthetic Biology, 2012, 1(1): 14-21. |
| 17 | MONTAGUE T G, CRUZ J M, GAGNON J A, et al. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing[J]. Nucleic Acids Research, 2014, 42(W1): W401-W407. |
| 18 | 唐婷, 付立豪, 郭二鹏, 等. 自动化合成生物技术与工程化设施平台[J]. 科学通报, 2021, 66(3): 300-309. |
| TANG T, FU L H, GUO E P, et al. Automation in synthetic biology using biological foundries[J]. Chinese Science Bulletin, 2021, 66(3): 300-309. | |
| 19 | ARNOLD C. Cloud labs: Where robots do the research[J]. Nature, 2022, 606(7914): 612-613. |
| 20 | WANG Y, LIU Y, LIU J, et al. MACBETH: Multiplex automated Corynebacterium glutamicum base editing method[J]. Metabolic Engineering, 2018, 47: 200-210. |
| 21 | CÁRDENAS P, ESHERICK L Y, CHAMBONNIER G, et al. GeneTargeter: automated in silico design for genome editing in the malaria parasite, Plasmodium falciparum [J]. The CRISPR Journal, 2022, 5(1): 155-164. |
| 22 | KÄMPKE T, KIENINGER M, MECKLENBURG M. Efficient primer design algorithms[J]. Bioinformatics, 2001, 17(3): 214-225. |
| 23 | LINZ U, DELLING U, RÜBSAMEN-WAIGMANN H. Systematic studies on parameters influencing the performance of the polymerase chain reaction[J]. Journal of Clinical Chemistry and Clinical Biochemistry (Zeitschrift Fur Klinische Chemie Und Klinische Biochemie), 1990, 28(1): 5-13. |
| 24 | ABD-ELSALAM K A. Bioinformatic tools and guideline for PCR primer design[J]. African Journal of Biotechnology, 2003, 2(5): 91-95. |
| 63 | POTAPOV V, ONG J L, KUCERA R B, et al. Comprehensive profiling of four base overhang ligation fidelity by T4 DNA ligase and application to DNA assembly[J]. ACS Synthetic Biology, 2018, 7(11): 2665-2674. |
| 64 | CASINI A, CHANG F Y, ELUERE R, et al. A pressure test to make 10 molecules in 90 days: external evaluation of methods to engineer biology[J]. Journal of the American Chemical Society, 2018, 140(12): 4302-4316. |
| 65 | 李洋, 申晓林, 孙新晓, 等. CRISPR基因编辑技术在微生物合成生物学领域的研究进展[J]. 合成生物学, 2021, 2(1): 106-120. |
| LI Y, SHEN X L, SUN X X, et al. Advances of CRISPR gene editing in microbial synthetic biology[J]. Synthetic Biology Journal, 2021, 2(1): 106-120. | |
| 66 | DELTCHEVA E, CHYLINSKI K, SHARMA C M, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III[J]. Nature, 2011, 471(7340): 602-607. |
| 67 | GRAHAM D B, ROOT D E. Resources for the design of CRISPR gene editing experiments[J]. Genome Biology, 2015, 16: 260. |
| 68 | CHUAI G H, WANG Q-L, LIU Q. In silico meets in vivo: towards computational CRISPR-based sgRNA design[J]. Trends in Biotechnology, 2017, 35(1): 12-21. |
| 69 | 王远立, 啜国晖, 闫继芳, 等. 计算机辅助CRISPR向导RNA设计[J]. 生物工程学报, 2017, 33(10): 1744-1756. |
| WANG Y L, CHUAI G H, YAN J F, et al. In silico CRISPR-based sgRNA design[J]. Chinese Journal of Biotechnology, 2017, 33(10): 1744-1756. | |
| 70 | DOUDNA J A, CHARPENTIER E. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096. |
| 71 | DOENCH J G, FUSI N, SULLENDER M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9[J]. Nature Biotechnology, 2016, 34(2): 184-191. |
| 72 | XU H, XIAO T F, CHEN C H, et al. Sequence determinants of improved CRISPR sgRNA design[J]. Genome Research, 2015, 25(8): 1147-1157. |
| 25 | 张新宇, 高燕宁. PCR引物设计及软件使用技巧[J]. 生物信息学, 2004, 2(4): 15-18, 46. |
| ZHANG X Y, GAO Y N. To design PCR primers with oligo 6 and primer premier 5[J]. Bioinformatiocs, 2004, 2(4): 15-18, 46. | |
| 73 | WANG T, WEI J J, SABATINI D M, et al. Genetic screens in human cells using the CRISPR-Cas9 system[J]. Science, 2014, 343(6166): 80-84. |
| 74 | WU X B, SCOTT D A, KRIZ A J, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells[J]. Nature Biotechnology, 2014, 32(7): 670-676. |
| 75 | DOENCH J G, HARTENIAN E, GRAHAM D B, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation[J]. Nature Biotechnology, 2014, 32(12): 1262-1267. |
| 76 | CHARI R, MALI P, MOOSBURNER M, et al. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach[J]. Nature Methods, 2015, 12(9): 823-826. |
| 77 | HSU P D, SCOTT D A, WEINSTEIN J A, et al. DNA targeting specificity of RNA-guided Cas9 nucleases[J]. Nature Biotechnology, 2013, 31(9): 827-832. |
| 78 | KIM D, KIM S, KIM S, et al. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq[J]. Genome Research, 2016, 26(3): 406-415. |
| 79 | SINGH R, KUSCU C, QUINLAN A, et al. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction[J]. Nucleic Acids Research, 2015, 43(18): e118. |
| 80 | KIM D, KIM J S. DIG-seq: a genome-wide CRISPR off-target profiling method using chromatin DNA[J]. Genome Research, 2018, 28(12): 1894-1900. |
| 81 | MORGENS D W, WAINBERG M, BOYLE E A, et al. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens[J]. Nature Communications, 2017, 8: 15178. |
| 82 | AGUIRRE A J, MEYERS R M, WEIR B A, et al. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting[J]. Cancer Discovery, 2016, 6(8): 914-929. |
| 83 | JIANG W Y, BIKARD D, COX D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nature Biotechnology, 2013, 31(3): 233-239. |
| 84 | ZHANG Y L, GE X L, YANG F Y, et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells[J]. Scientific Reports, 2014, 4: 5405. |
| 85 | PATTANAYAK V, LIN S, GUILINGER J P, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity[J]. Nature Biotechnology, 2013, 31(9): 839-843. |
| 86 | CRADICK T J, FINE E J, ANTICO C J, et al. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity[J]. Nucleic Acids Research, 2013, 41(20): 9584-9592. |
| 87 | LANGMEAD B, TRAPNELL C, POP M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome[J]. Genome Biology, 2009, 10(3): R25. |
| 88 | LI H, DURBIN R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25(14): 1754-1760. |
| 89 | JIANG H, WONG W H. SeqMap: mapping massive amount of oligonucleotides to the genome[J]. Bioinformatics, 2008, 24(20): 2395-2396. |
| 90 | BAE S S, PARK J, KIM J S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases[J]. Bioinformatics, 2014, 30(10): 1473-1475. |
| 91 | XIAO A, CHENG Z C, KONG L, et al. CasOT: a genome-wide Cas9/gRNA off-target searching tool[J]. Bioinformatics, 2014, 30(8): 1180-1182. |
| 92 | CANCELLIERI S, CANVER M C, BOMBIERI N, et al. CRISPRitz: rapid, high-throughput and variant-aware in silico off-target site identification for CRISPR genome editing[J]. Bioinformatics, 2020, 36(7): 2001-2008. |
| 93 | STEMMER M, THUMBERGER T, DEL SOL KEYER M, et al. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool[J]. PLoS One, 2017, 12(4): e0124633. |
| 94 | HAEUSSLER M, SCHÖNIG K, ECKERT H, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR[J]. Genome Biology, 2016, 17(1): 148. |
| 95 | XIE S S, SHEN B, ZHANG C B, et al. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites[J]. PLoS One, 2014, 9(6): e100448. |
| 96 | JACQUIN A L S, ODOM D T, LUKK M. Crisflash: open-source software to generate CRISPR guide RNAs against genomes annotated with individual variation[J]. Bioinformatics, 2019, 35(17): 3146-3147. |
| 97 | MCKENNA A, SHENDURE J. FlashFry: a fast and flexible tool for large-scale CRISPR target design[J]. BMC Biology, 2018, 16(1): 74. |
| 98 | HEIGWER F, KERR G, BOUTROS M. E-CRISP: fast CRISPR target site identification[J]. Nature Methods, 2014, 11(2): 122-123. |
| 99 | LIU H L, WEI Z, DOMINGUEZ A, et al. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation[J]. Bioinformatics, 2015, 31(22): 3676-3678. |
| 100 | PENG D, TARLETON R. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens[J]. Microbial Genomics, 2015, 1(4): e000033. |
| 101 | CONCORDET J P, HAEUSSLER M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens[J]. Nucleic Acids Research, 2018, 46(W1): W242-W245. |
| 102 | LABUN K, MONTAGUE T G, GAGNON J A, et al. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering[J]. Nucleic Acids Research, 2016, 44(W1): W272-W276. |
| 103 | LABUN K, MONTAGUE T G, KRAUSE M, et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing[J]. Nucleic Acids Research, 2019, 47(W1): W171-W174. |
| 104 | PULIDO-QUETGLAS C, APARICIO-PRAT E, ARNAN C, et al. Scalable design of paired CRISPR guide RNAs for genomic deletion[J]. PLoS Computational Biology, 2017, 13(3): e1005341. |
| 105 | LEI Y, LU L, LIU H Y, et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants[J]. Molecular Plant, 2014, 7(9): 1494-1496. |
| 106 | MORENO-MATEOS M A, VEJNAR C E, BEAUDOIN J D, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo [J]. Nature Methods, 2015, 12(10): 982-988. |
| 107 | WONG N, LIU W J, WANG X W. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system[J]. Genome Biology, 2015, 16: 218. |
| 108 | ABADI S, YAN W X, AMAR D, et al. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action[J]. PLoS Computational Biology, 2017, 13(10): e1005807. |
| 109 | LISTGARTEN J, WEINSTEIN M, KLEINSTIVER B P, et al. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs[J]. Nature Biomedical Engineering, 2018, 2(1): 38-47. |
| 110 | KONSTANTAKOS V, NENTIDIS A, KRITHARA A, et al. CRISPR-Cas9 gRNA efficiency prediction: an overview of predictive tools and the role of deep learning[J]. Nucleic Acids Research, 2022, 50(7): 3616-3637. |
| 111 | WANG D Q, ZHANG C D, WANG B, et al. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning[J]. Nature Communications, 2019, 10: 4284. |
| 112 | HWANG G H, PARK J, LIM K, et al. Web-based design and analysis tools for CRISPR base editing[J]. BMC Bioinformatics, 2018, 19(1): 542. |
| 113 | DANDAGE R, DESPRÉS P C, YACHIE N, et al. Beditor: a computational workflow for designing libraries of guide RNAs for CRISPR-mediated base editing[J]. Genetics, 2019, 212(2): 377-385. |
| 114 | WANG Y, LIU Y, LI J W, et al. Expanding targeting scope, editing window, and base transition capability of base editing in Corynebacterium glutamicum [J]. Biotechnology and Bioengineering, 2019, 116(11): 3016-3029. |
| 115 | MANS R, VAN ROSSUM H M, WIJSMAN M, et al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae [J]. FEMS Yeast Research, 2015, 15(2): fov004. |
| 116 | YANG Y, MAO Y F, LIU Y, et al. GEDpm-cg: genome editing automated design platform for point mutation construction in Corynebacterium glutamicum [J]. Frontiers in Bioengineering and Biotechnology, 2021, 9: 768289. |
| 117 | TONG Y J, JØRGENSEN T S, WHITFORD C M, et al. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing[J]. Nature Communications, 2021, 12: 5206. |
| 118 | YANG X, YUAN Q Q, LUO H, et al. Systematic design and in vitro validation of novel one-carbon assimilation pathways[J]. Metabolic Engineering, 2019, 56: 142-153. |
| 119 | LIAO X P, MA H W, TANG Y J. Artificial intelligence: a solution to involution of design-build-test-learn cycle[J]. Current Opinion in Biotechnology, 2022, 75: 102712. |
| 120 | SABZEVARI M, SZEDMAK S, PENTTILÄ M, et al. Strain design optimization using reinforcement learning[J]. PLoS Computational Biology, 2022, 18(6): e1010177. |
| 121 | WANG Y, WANG H C, WEI L, et al. Synthetic promoter design in Escherichia coli based on a deep generative network[J]. Nucleic Acids Research, 2020, 48(12): 6403-6412. |
| 122 | SALIS H M, MIRSKY E A, VOIGT C A. Automated design of synthetic ribosome binding sites to control protein expression[J]. Nature Biotechnology, 2009, 27(10): 946-950. |
| [1] | CHEN Yongcan, SI Tong, ZHANG Jianzhi. Applications of automated synthetic biotechnology in DNA assembly and microbial chassis manipulation [J]. Synthetic Biology Journal, 2023, 4(5): 857-876. |
| [2] | HU Zhehui, XU Juan, BIAN Guangkai. Application of automated high-throughput technology in natural product biosynthesis [J]. Synthetic Biology Journal, 2023, 4(5): 932-946. |
| [3] | LU Hui, ZHANG Fangli, HUANG Lei. Establishment of iBioFoundry for synthetic biology applications [J]. Synthetic Biology Journal, 2023, 4(5): 877-891. |
| [4] | ZHAO Guomiao, YANG Xin, ZHANG Yuan, WANG Jing, TAN Jian, WEI Chao, ZHOU Nana, LI Fan, WANG Xiaoyan. Biofoundry and its industrial application [J]. Synthetic Biology Journal, 2023, 4(5): 892-903. |
| [5] | Xiaolong TENG, Shuobo SHI. Optimization and development of CRISPR/Cas9 systems for genome editing [J]. Synthetic Biology Journal, 2023, 4(1): 67-85. |
| [6] | Chanjuan JIANG, Tianqi CUI, Hongluan SUN, Nianzhi JIAO, Jun FU, Youming ZHANG, Hailong WANG. Efficient capture and assembly of AT-rich genomic fragments using ExoCET-BAC strategy [J]. Synthetic Biology Journal, 2022, 3(1): 238-251. |
| [7] | Qingzhuo WANG, Ping SONG, He HUANG. Synthetic biotechnology drives the development of natural eukaryotic lipid cell factories [J]. Synthetic Biology Journal, 2021, 2(6): 920-941. |
| [8] | Qian LIU, Jingen LI, Chenyang ZHANG, Fangya LI, Chaoguang TIAN. Research progress of genome editing technologies for industrial filamentous fungi [J]. Synthetic Biology Journal, 2021, 2(2): 256-273. |
| [9] | Kai PENG, Xiaoyun LU, Jian CHENG, Ying LIU, Huifeng JIANG, Xiaoxian GUO. Advances in technologies for de novo DNA synthesis, assembly and error correction [J]. Synthetic Biology Journal, 2020, 1(6): 697-708. |
| [10] | Yong YU, Xinna ZHU, Xueli ZHANG. Construction and application of microbial cell factories for production of bulk chemicals [J]. Synthetic Biology Journal, 2020, 1(6): 674-684. |
| [11] | Hui WANG, Junbiao DAI, Zhouqing LUO. Reading, editing, and writing techniques for genome research [J]. Synthetic Biology Journal, 2020, 1(5): 503-515. |
| [12] | Zhongzheng CAO, Xinyi ZHANG, Yiyuan XU, Zhuo ZHOU, Wensheng WEI. Genome editing technology and its applications in synthetic biology [J]. Synthetic Biology Journal, 2020, 1(4): 413-426. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||