Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (6): 697-708.DOI: 10.12211/2096-8280.2020-034
• Invited Review • Previous Articles Next Articles
Advances in technologies for de novo DNA synthesis, assembly and error correction
PENG Kai1,2, LU Xiaoyun1, CHENG Jian1, LIU Ying1, JIANG Huifeng1, GUO Xiaoxian1
- 1.Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,Tianjin 300308,China
2.School of Biological Engineering,Tianjin University of Science and Technology,Tianjin 300457,China
-
Received:2020-03-23Revised:2020-10-22Online:2021-01-15Published:2020-12-31 -
Contact:JIANG Huifeng, GUO Xiaoxian
DNA合成、组装与纠错技术研究进展
彭凯1,2, 逯晓云1, 程健1, 刘莹1, 江会锋1, 郭晓贤1
- 1.中国科学院天津工业生物技术研究所系统微生物工程重点实验室,天津 300308
2.天津科技大学生物工程学院,天津 300457
-
通讯作者:江会锋,郭晓贤 -
作者简介:彭凯(1995—),男,硕士研究生。主要研究方向为DNA纠错。E-mail: pengk@tib.cas.cn
江会锋(1981—),男,博士,研究员。主要研究方向为代谢合成生物学。E-mail: jiang_hf@tib.cas.cn
郭晓贤(1982—),男,博士,副研究员。主要研究方向为酶法DNA合成。E-mail: guoxx@tib.cas.cn -
基金资助:国家重点研发计划(2020YFC1316400);天津市合成生物技术创新能力提升行动(TSBICIP-KJGG-007-01)
CLC Number:
Cite this article
PENG Kai, LU Xiaoyun, CHENG Jian, LIU Ying, JIANG Huifeng, GUO Xiaoxian. Advances in technologies for de novo DNA synthesis, assembly and error correction[J]. Synthetic Biology Journal, 2020, 1(6): 697-708.
彭凯, 逯晓云, 程健, 刘莹, 江会锋, 郭晓贤. DNA合成、组装与纠错技术研究进展[J]. 合成生物学, 2020, 1(6): 697-708.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-034
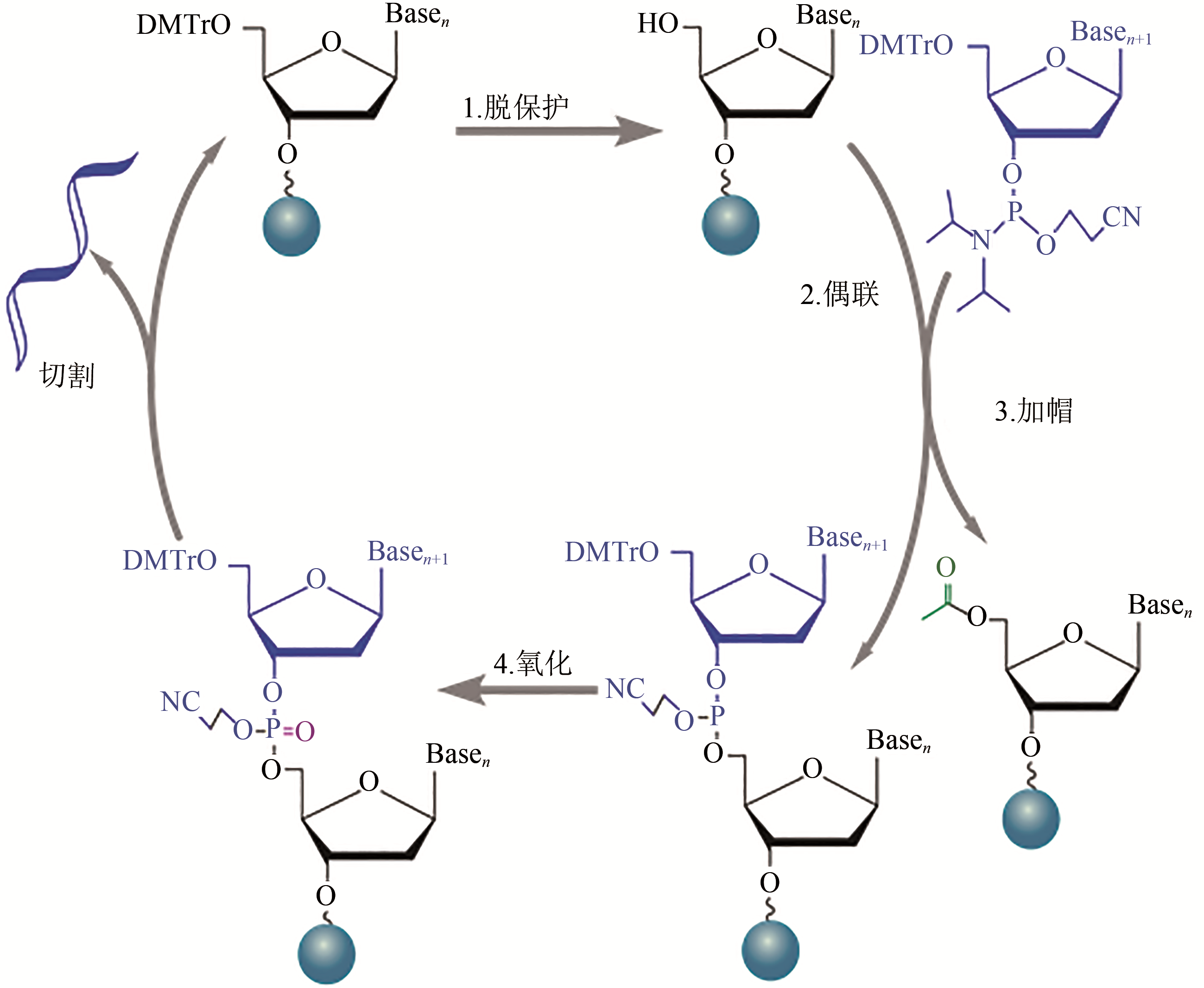
Fig. 1 Reaction cycle for solid-phase phosphoramidite synthesis of oligonucleotide[1—Deprotection. The dimethoxytrityl (DMT) ether capping group on the 5’ end of the previous nucleotide is removed to generate the 5’ end hydroxyl group on the growing chain. 2—Coupling. The desired phosphoramidite monomer couples to the chain via the 5’ end hydroxyl group generated in the deprotection step. 3—Capping. Uncoupled 5’ end hydroxyl group is blocked by acylating molecules to prevent further reactions. 4—Oxidation. An oxidizer agent is used to converte phosphite triester linkage to a more stable phosphotriester bond prior to the start of the next cycle. Cleavage: final oligonucleotides are cleaved from their solid support at the end of synthesis]
| 1 | GIBSON D G. Programming biological operating systems: genome design, assembly and activation[J]. Nature Methods, 2014, 11(5): 521-526. |
| 2 | CARROLL D. Genome engineering with targetable nucleases[J]. Annual Review Biochemistry, 2014, 83: 409-439. |
| 3 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 4 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age[J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 5 | ENDY D. Foundations for engineering biology[J]. Nature, 2005, 438(7067): 449-453. |
| 6 | MA S, TANG N, TIAN J. DNA synthesis, assembly and applications in synthetic biology[J]. Current Opinion in Chemical Biology, 2012, 16(3/4): 260-267. |
| 7 | CARUTHERS M H. The chemical synthesis of DNA/RNA: our gift to science[J]. Journal of Biological Chemistry, 2013, 288(2): 1420-1427. |
| 8 | PALLUK S, ARLOW D H, DE ROND T, et al. De novo DNA synthesis using polymerase-nucleotide conjugates[J]. Nature Biotechnology, 2018, 36(7): 645-650. |
| 9 | CZAR M J, ANDERSON J C, BADER J S, et al. Gene synthesis demystified[J]. Trends in Biotechnology, 2009, 27(2): 63-72. |
| 10 | MA S, SAAEM I, TIAN J. Error correction in gene synthesis technology[J]. Trends in Biotechnology, 2012, 30(3): 147-154. |
| 11 | BEAUCAGE S L, CARUTHERS M H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis[J]. Tetrahedron Letters, 1981, 22(20): 1859-1862. |
| 12 | TIAN J, MA K, SAAEM I. Advancing high-throughput gene synthesis technology[J]. Molecular Biosystems, 2009, 5(7): 714-722. |
| 13 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications[J]. Nature Methods, 2014, 11(5): 499-507. |
| 14 | LEPROUST E M, PECK B J, SPIRIN K, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process[J]. Nucleic Acids Research, 2010, 38(8): 2522-2540. |
| 15 | JENSEN Michael, ROBERTS Lester, JOHNSON Andrew, et al. Next generation 1536-well oligonucleotide synthesizer with on-the-fly dispense[J]. Journal of biotechnology, 2014, 171: 76-81. |
| 16 | FODOR S P, READ J L, PIRRUNG M C,et al. Light-directed, spatially addressable parallel chemical synthesis[J]. Science, 1991, 251(4995): 767-773. |
| 17 | BARONE A D, BEECHER J E, BURY P A, et al. Photolithographic synthesis of high-density oligonucleotide probe arrays[J]. Nucleosides Nucleic Acids, 2001, 20(4-7): 525-531. |
| 18 | SINGH-GASSON S, GREEN R D, YUE Y, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array[J]. Nature Biotechnology, 1999, 17(10): 974-978. |
| 19 | GAO X, LEPROUST E, ZHANG H, et al. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids[J]. Nucleic Acids Research, 2001, 29(22): 4744-4750. |
| 20 | AGBAVWE C, KIM C, HONG D, et al. Efficiency, error and yield in light-directed maskless synthesis of DNA microarrays[J]. Journal of Nanobiotechnology, 2011, 9: 57. |
| 21 | ROTH Kristian M, PEYVAN Kia, SCHWARZKOPF Kevin R, et al. Electrochemical detection of short DNA oligomer hybridization using the CombiMatrix ElectraSense microarray reader[J]. Electroanalysis, 2006, 18(19‐20): 1982-1988. |
| 22 | HUGHES T R, MAO M, JONES A R, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer[J]. Nature Biotechnology, 2001, 19(4): 342-347. |
| 23 | LIPSHUTZ R J, FODOR S P, GINGERAS T R, et al. High density synthetic oligonucleotide arrays[J]. Nature Genetics, 1999, 21(S1): 20-24. |
| 24 | MINHAZ UD-DEAN S M. A theoretical model for template-free synthesis of long DNA sequence[J]. Systems and Synthetic Biology, 2008, 2(3/4): 67-73. |
| 25 | MACKEY J K, GILHAM P T. New approach to the synthesis of polyribonucleotides of defined sequence[J]. Nature, 1971, 233(5321): 551-553. |
| 26 | GILLAM S, WATERMAN K, DOEL M, et al. Enzymatic synthesis of deoxyribo-oligonucleotides of defined sequence. Deoxyribo-oligonucleotide synthesis[J]. Nucleic Acids Research, 1974, 1(12): 1649-1664. |
| 27 | ENGLAND T E, UHLENBECK O C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase[J]. Biochemistry, 1978, 17(11): 2069-2076. |
| 28 | SCHMITZ Carole, REETZ Manfred T. Solid-phase enzymatic synthesis of oligonucleotides[J]. Organic Letters, 1999, 1(11): 1729-1731. |
| 29 | JENSEN M A, DAVIS R W. Template-independent enzymatic oligonucleotide synthesis (TiEOS): its history, prospects, and challenges[J]. Biochemistry, 2018, 57(12): 1821-1832. |
| 30 | BOLLUM F J. Thermal conversion of nonpriming deoxyribonucleic acid to primer [J]. The Journal of Biological Chemistry, 1959, 234(10): 2733-2734. |
| 31 | BOLLUM F J. Oligodeoxyribonucleotide-primed reactions catalyzed by calf thymus polymerase[J]. Journal of Biological Chemistry, 1962, 237: 1945-1949. |
| 32 | SCHOTT H, SCHRADE H. Single-step elongation of oligodeoxynucleotides using terminal deoxynucleotidyl transferase[J]. European Journal of Biochemistry, 1984, 143(3): 613-620. |
| 33 | TJONG V, YU H, HUCKNALL A, et al. Amplified on-chip fluorescence detection of DNA hybridization by surface-initiated enzymatic polymerization[J]. Analytical Chemistry, 2011, 83(13): 5153-5159. |
| 34 | MOTEA E A, BERDIS A J. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase[J]. Biochimica et Biophysica Acta(BBA) - Proteins and Proteomics, 2010, 1804(5): 1151-1166. |
| 35 | WU J, ZHANG S, MENG Q, et al. 3'-O-modified nucleotides as reversible terminators for pyrosequencing[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(42): 16462-16467. |
| 36 | KONG D S, CARR P A, CHEN L, et al. Parallel gene synthesis in a microfluidic device[J]. Nucleic Acids Research, 2007, 35(8): e61. |
| 37 | TIAN J, GONG H, SHENG N, et al. Accurate multiplex gene synthesis from programmable DNA microchips[J]. Nature, 2004, 432(7020): 1050-1054. |
| 38 | QUAN J, SAAEM I, TANG N, et al. Parallel on-chip gene synthesis and application to optimization of protein expression[J]. Nature Biotechnology, 2011, 29(5): 449-452. |
| 39 | ENGLER C, KANDZIA R, MARILLONNET S. A one pot, one step, precision cloning method with high throughput capability[J]. PLoS One, 2008, 3(11): e3647. |
| 40 | LI M Z, ELLEDGE S J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC[J]. Nature Methods, 2007, 4(3): 251-256. |
| 41 | ZHANG Y, WERLING U, EDELMANN W. SLiCE: a novel bacterial cell extract-based DNA cloning method[J]. Nucleic Acids Research, 2012, 40(8): e55. |
| 42 | Stefan de KOK, STANTON Leslie H, SLABY Todd, et al. Rapid and reliable DNA assembly via ligase cycling reaction[J]. ACS Synthetic Biology, 2014, 3(2): 97-106. |
| 43 | QUAN J, TIAN J. Circular polymerase extension cloning of complex gene libraries and pathways[J]. PLoS One, 2009, 4(7): e6441. |
| 44 | GIBSON D G, YOUNG L, CHUANG R Y, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nature Methods, 2009, 6(5): 343-345. |
| 45 | JUHAS M, AJIOKA J W. High molecular weight DNA assembly in vivo for synthetic biology applications[J]. Critical Reviews in Biotechnology, 2017, 37(3): 277-286. |
| 46 | SHIZUYA H, BIRREN B, KIM U J, et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector[J]. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(18): 8794-8797. |
| 47 | KANEKO S, TSUGE K, TAKEUCHI T, et al. Conversion of sub-megasized DNA to desired structures using a novel Bacillus subtilis genome vector[J]. Nucleic Acids Research, 2003, 31(18): e112. |
| 48 | BURKE D T, CARLE G F, OLSON M V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors[J]. Science, 1987, 236(4803): 806-812. |
| 49 | OGAWA T, IWATA T, KANEKO S, et al. An inducible recA expression Bacillus subtilis genome vector for stable manipulation of large DNA fragments[J]. Biotechnology & Applied Microbiology Genomics, 2015, 16(1): 209. |
| 50 | GIBSON D G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides[J]. Nucleic Acids Research, 2009, 37(20): 6984-6990. |
| 51 | LIN Q, JIA B, MITCHELL L A, et al. RADOM, an efficient in vivo method for assembling designed DNA fragments up to 10 kb long in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2015, 4(3): 213-220. |
| 52 | JAKOCIUNAS T, RAJKUMAR A S, ZHANG J, et al. CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2015, 4(11): 1226-1234. |
| 53 | GIBSON D G, BENDERS G A, AXELROD K C, et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(51): 20404-20409. |
| 54 | SHAO Y, LU N, WU Z, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 55 | LUBOCK N B, ZHANG D, SIDORE A M, et al. A systematic comparison of error correction enzymes by next-generation sequencing[J]. Nucleic Acids Research, 2017, 45(15): 9206-9217. |
| 56 | SINHA N D, JUNG K E. Analysis and purification of synthetic nucleic acids using HPLC[J]. Current Protocols in Nucleic Acid Chemistry, 2015. DOI: 10.1002/0471142700.nc1005s61 . |
| 57 | ELLINGTON A, POLLARD J D, Jr. Introduction to the synthesis and purification of oligonucleotides[J]. Current Protocols in Nucleic Acid Chemistry, 2000. DOI: 10.1002/0471142700. nca03cs00 . |
| 58 | BOROVKOV A Y, LOSKUTOV A V, ROBIDA M D, et al. High-quality gene assembly directly from unpurified mixtures of microarray-synthesized oligonucleotides[J]. Nucleic Acids Research, 2010, 38(19): e180. |
| 59 | MATZAS M, STAHLER P F, KEFER N, et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing[J]. Nature Biotechnology, 2010, 28(12): 1291-1294. |
| 60 | SANCAR A, LINDSEY-BOLTZ L A, UNSAL-KACMAZ K, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints[J]. Annual Review of Biochemistry, 2004, 73: 39-85. |
| 61 | JIRICNY J. The multifaceted mismatch-repair system[J]. Nature Reviews Molecular Cell Biology, 2006, 7(5): 335-346. |
| 62 | LEE J B, CHO W K, PARK J, et al. Single-molecule views of MutS on mismatched DNA[J]. DNA Repair (Amst), 2014, 20: 82-93. |
| 63 | KUNKEL T A, ERIE D A. DNA mismatch repair[J]. Annual Review of Biochemistry, 2005, 74: 681-710. |
| 64 | CARR P A, PARK J S, LEE Y J, et al. Protein-mediated error correction for de novo DNA synthesis[J]. Nucleic Acids Research, 2004, 32(20): e162. |
| 65 | WAN W, LI L, XU Q, et al. Error removal in microchip-synthesized DNA using immobilized MutS[J]. Nucleic Acids Research, 2014, 42(12): e102. |
| 66 | ZHANG J, WANG Y, CHAI B, et al. Efficient and low-cost error removal in DNA synthesis by a high-durability MutS[J]. ACS Synthetic Biology, 2020, 9(4): 940-952. |
| 67 | BINKOWSKI B F, RICHMOND K E, KAYSEN J, et al. Correcting errors in synthetic DNA through consensus shuffling[J]. Nucleic Acids Research, 2005, 33(6): e55. |
| 68 | TILL B J, BURTNER C, COMAI L, et al. Mismatch cleavage by single-strand specific nucleases[J]. Nucleic Acids Research, 2004, 32(8): 2632-2641. |
| 69 | FUHRMANN M. Removal of mismatched bases from synthetic genes by enzymatic mismatch cleavage[J]. Nucleic Acids Research, 2005, 33(6): e58. |
| 70 | DESAI N A, SHANKAR V. Single-strand-specific nucleases[J]. FEMS Microbiology Reviews, 2003, 26(5): 457-491. |
| 71 | SEQUEIRA A F, GUERREIRO C I, VINCENTELLI R, et al. T7 Endonuclease I mediates error correction in artificial gene synthesis[J]. Molecular Biotechnology, 2016, 58(8-9): 573-584. |
| 72 | BANG D, CHURCH G M. Gene synthesis by circular assembly amplification[J]. Nature Methods, 2008, 5(1): 37-39. |
| 73 | BABON J J, MCKENZIE M, COTTON R G. Mutation detection using fluorescent enzyme mismatch cleavage with T4 endonuclease Ⅶ[J]. Electrophoresis, 1999, 20(6): 1162-1170. |
| 74 | YEUNG A T, HATTANGADI D, BLAKESLEY L, et al. Enzymatic mutation detection technologies[J]. Biotechniques, 2005, 38(5): 749-758. |
| 75 | OLEYKOWSKI C A, BRONSON MULLINS C R, GODWIN A K, et al. Mutation detection using a novel plant endonuclease[J]. Nucleic Acids Research, 1998, 26(20): 4597-4602. |
| 76 | YANG B, WEN X, KODALI N S, et al. Purification, cloning, and characterization of the CEL I nuclease[J]. Biochemistry, 2000, 39(13): 3533-3541. |
| 77 | SAAEM I, MA S, QUAN J, et al. Error correction of microchip synthesized genes using Surveyor nuclease[J]. Nucleic Acids Research, 2012, 40(3): e23. |
| 78 | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synⅡ, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355(6329) : eaaf4791. |
| 79 | WU Y, LI B Z, ZHAO M,et al. Bug mapping and fitness testing of chemically synthesized chromosome Ⅹ[J]. Science, 2017, 355(6329) : eaaf4706. |
| 80 | XIE Z X, LI B Z, MITCHELL L A, et al. ‘Perfect’ designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329) : eaaf4704. |
| 81 | ZHANG W, ZHAO G, LUO Z, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355(6329) : eaaf3981. |
| 82 | RICHARDSON S M, MITCHELL L A, STRACQUADANIO G., et al. Design of a synthetic yeast genome[J]. Science, 2017, 355(6329): 1040-1044. |
| 83 | SEEMAN Nadrian C, SLEIMAN Hanadi F. DNA nanotechnology[J]. Nature Reviews Materials, 2017, 3(1) :17068. |
| 84 | GOLDMAN N, BERTONE P, CHEN S, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA[J]. Nature, 2013, 494(7435): 77-80. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [7] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [8] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [9] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [10] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [11] | ZHANG Xuanliang, LI Qingting, WANG Fei. Data writing in DNA storage systems [J]. Synthetic Biology Journal, 2024, 5(5): 1125-1141. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||