Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (5): 966-979.DOI: 10.12211/2096-8280.2023-033
• Invited Review • Previous Articles Next Articles
Research and application progress of microdroplets high throughput screening methods
QIN Weitong, YANG Guangyu
- State Key Laboratory of Microbial Metabolism,School of Life Science and Technology,Shanghai Jiaotong University,Shanghai 200240,China
-
Received:2023-04-24Revised:2023-06-20Online:2023-11-15Published:2023-10-31 -
Contact:YANG Guangyu
微液滴高通量筛选方法的研究与应用进展
秦伟彤, 杨广宇
- 上海交通大学生命科学技术学院,微生物代谢国家重点实验室,上海 200240
-
通讯作者:杨广宇 -
作者简介:秦伟彤 (1992—),女,博士。研究方向为酶的定向进化、微液滴超高通量筛选方法的建立。E-mail:qinweitong122@163.com杨广宇 (1980—),男,研究员,博士生导师。研究方向为酶分子改造、超高通量筛选方法的建立;酶分子机制解析;体外合成生物学研究。E-mail:yanggy@sjtu.edu.cn -
基金资助:国家重点研发计划(2018YFE0200501);国家自然科学基金(32030063);广东省重点领域研发计划(2022B1111050001);天津市合成生物技术创新能力提升行动;上海交通大学特区计划(21TQ1400210)
CLC Number:
Cite this article
QIN Weitong, YANG Guangyu. Research and application progress of microdroplets high throughput screening methods[J]. Synthetic Biology Journal, 2023, 4(5): 966-979.
秦伟彤, 杨广宇. 微液滴高通量筛选方法的研究与应用进展[J]. 合成生物学, 2023, 4(5): 966-979.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-033

Fig. 2 The principle of labeled droplet sorting technologyFADS—Fluorescence activated droplet sorting technology; AADS—Absorbance activated droplet separation technology; S—Substrate; P—Product; E: Enzyme molecules; Ex—Emission spectrum; Em—Absorption spectrum
| 分选方法 | 最高分选效率 | 最高分选灵敏度 | 最小液滴体积 | 优点 | 缺点 |
|---|---|---|---|---|---|
| FADS | 5 kHz[ | 2.5 nmol/L[ | 2 pL[ | 检测灵敏度高、分选速度快、平台发展成熟 | 大部分检测靶标缺乏适合的荧光耦联方法 |
| AADS | 1 kHz[ | 10 μmol/L[ | 100 pL[ | 普适性较FADS高 | 检测灵敏度待提高 |
| MADS | 35 Hz[ | 5 μmol/L[ | 0.8 nL[ | 无损伤、普适性高 | 分选速度慢、灵敏度待提高 |
| RADS | 4.3 Hz[ | 50 μmol/L[ | 65 pL[ | 无损伤、普适性高 | 更适用于较大的细胞 |
| NMR-ADS | — | 1 mmol/L[ | 130 pL[ | 无损伤、提供信息广泛 | NMR与液滴分选系统的整合较困难、检测灵敏度低 |
| IBDS | 10 Hz[ | — | 35 pL[ | 无损伤 | 适用范围窄、分选速度慢 |
| EADS | 10 Hz[ | 1 μmol/L[ | 30 nL[ | 无损伤 | 适用范围窄、分选速度慢 |
Table 1 Comparison of different microfluidic sorting equipment
| 分选方法 | 最高分选效率 | 最高分选灵敏度 | 最小液滴体积 | 优点 | 缺点 |
|---|---|---|---|---|---|
| FADS | 5 kHz[ | 2.5 nmol/L[ | 2 pL[ | 检测灵敏度高、分选速度快、平台发展成熟 | 大部分检测靶标缺乏适合的荧光耦联方法 |
| AADS | 1 kHz[ | 10 μmol/L[ | 100 pL[ | 普适性较FADS高 | 检测灵敏度待提高 |
| MADS | 35 Hz[ | 5 μmol/L[ | 0.8 nL[ | 无损伤、普适性高 | 分选速度慢、灵敏度待提高 |
| RADS | 4.3 Hz[ | 50 μmol/L[ | 65 pL[ | 无损伤、普适性高 | 更适用于较大的细胞 |
| NMR-ADS | — | 1 mmol/L[ | 130 pL[ | 无损伤、提供信息广泛 | NMR与液滴分选系统的整合较困难、检测灵敏度低 |
| IBDS | 10 Hz[ | — | 35 pL[ | 无损伤 | 适用范围窄、分选速度慢 |
| EADS | 10 Hz[ | 1 μmol/L[ | 30 nL[ | 无损伤 | 适用范围窄、分选速度慢 |
| 发表时间 | 分选系统 | 目标酶 | 分选结果 | 参考文献 |
|---|---|---|---|---|
| 2018 | FADS | 酯酶 | 对S-布洛芬的对映选择性提高600倍 | [ |
| 2019 | FADS | 硫酸酯酶 | Kcat/Km值提高30倍 | [ |
| 2019 | FADS | 纤维素酶 | 筛选出产量提升46%的高纤维素酶菌株 | [ |
| 2020 | FADS | 葡萄糖氧化酶 | Kcat值比野生型高2.1倍 | [ |
| 2020 | AADS | 胺脱氢酶 | 转化率提高3.3倍 | [ |
| 2022 | FADS | α-淀粉酶 | 产量提升50%的地衣芽孢杆菌突变株 | [ |
| 2023 | FADS | 二乙酰壳二糖脱乙酰酶 | 催化效率提高1.8倍 | [ |
| 2022 | FADS | 塑料降解酶 | 2株可降解塑料的菌株 | [ |
| 2022 | FADS | 产鼠李糖脂的微生物 | 产量提升54%~208%的菌株 | [ |
| 2022 | AADS | 葡萄糖脱氢酶 | 催化速度和效率提升10倍以上 | [ |
Table 2 Cases of successful application of microfluidic sorting devices in the past five years
| 发表时间 | 分选系统 | 目标酶 | 分选结果 | 参考文献 |
|---|---|---|---|---|
| 2018 | FADS | 酯酶 | 对S-布洛芬的对映选择性提高600倍 | [ |
| 2019 | FADS | 硫酸酯酶 | Kcat/Km值提高30倍 | [ |
| 2019 | FADS | 纤维素酶 | 筛选出产量提升46%的高纤维素酶菌株 | [ |
| 2020 | FADS | 葡萄糖氧化酶 | Kcat值比野生型高2.1倍 | [ |
| 2020 | AADS | 胺脱氢酶 | 转化率提高3.3倍 | [ |
| 2022 | FADS | α-淀粉酶 | 产量提升50%的地衣芽孢杆菌突变株 | [ |
| 2023 | FADS | 二乙酰壳二糖脱乙酰酶 | 催化效率提高1.8倍 | [ |
| 2022 | FADS | 塑料降解酶 | 2株可降解塑料的菌株 | [ |
| 2022 | FADS | 产鼠李糖脂的微生物 | 产量提升54%~208%的菌株 | [ |
| 2022 | AADS | 葡萄糖脱氢酶 | 催化速度和效率提升10倍以上 | [ |
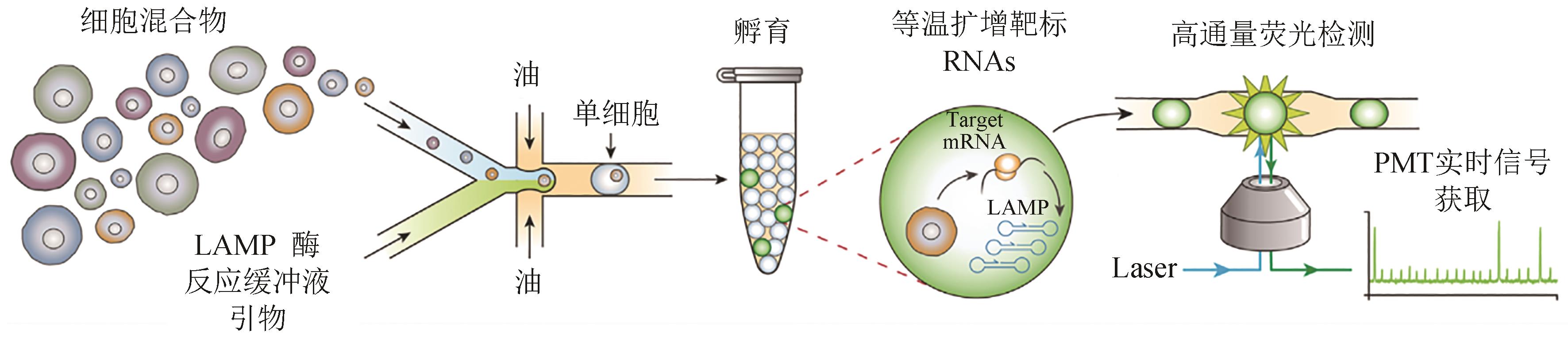
Fig. 4 Schematic of the SNAPD workflow[96](Single cells are encapsulated into microdroplets with assay reagents, collected and incubated offline, and the fluorescence of each droplet is subsequently measured to indicate amplification of target RNAs)
| 1 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 2 | XIONG W, LIU B, SHEN Y J, et al. Protein engineering design from directed evolution to de novo synthesis[J]. Biochemical Engineering Journal, 2021, 174: 108096. |
| 3 | LONGWELL C K, LABANIEH L, COCHRAN J R. High-throughput screening technologies for enzyme engineering[J]. Current Opinion in Biotechnology, 2017, 48: 196-202. |
| 4 | MACARRON R, BANKS M N, BOJANIC D, et al. Impact of high-throughput screening in biomedical research[J]. Nature Reviews Drug Discovery, 2011, 10(3): 188-195. |
| 5 | QIN W T, LI L, YANG F, et al. High-throughput iSpinach fluorescent aptamer-based real-time monitoring of in vitro transcription[J].Bioresources and Bioprocessing, 2022, 9: 112. |
| 6 | LLOYD M D. High-throughput screening for the discovery of enzyme inhibitors[J]. Journal of Medicinal Chemistry, 2020, 63(19): 10742-10772. |
| 7 | SARNAIK A, LIU A, NIELSEN D, et al. High-throughput screening for efficient microbial biotechnology[J]. Current Opinion in Biotechnology, 2020, 64: 141-150. |
| 8 | CAEN O, SCHÜTZ S, JAMMALAMADAKA M S S, et al. High-throughput multiplexed fluorescence-activated droplet sorting[J]. Microsystems & Nanoengineering, 2018, 4: 33. |
| 9 | POTYRAILO R, RAJAN K, STOEWE K, et al. Combinatorial and high-throughput screening of materials libraries: review of state of the art[J]. ACS Combinatorial Science, 2011, 13(6): 579-633. |
| 10 | CHIN C D, LINDER V, SIA S K. Lab-on-a-chip devices for global health: past studies and future opportunities[J]. Lab on a Chip, 2007, 7(1): 41-57. |
| 11 | AZIZIPOUR N, AVAZPOUR R, ROSENZWEIG D H, et al. Evolution of biochip technology: a review from lab-on-a-chip to organ-on-a-chip[J]. Micromachines, 2020, 11(6): 599. |
| 12 | BARET J C, MILLER O J, TALY V, et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity[J]. Lab on a Chip, 2009, 9(13): 1850-1858. |
| 13 | TU R, ZHANG Y, HUA E B, et al. Droplet-based microfluidic platform for high-throughput screening of Streptomyces[J]. Communications Biology, 2021, 4: 647. |
| 14 | SJOSTROM S L, BAI Y P, HUANG M T, et al. High-throughput screening for industrial enzyme production hosts by droplet microfluidics[J]. Lab on a Chip, 2014, 14(4): 806-813. |
| 15 | FU X Z, ZHANG Y Y, XU Q, et al. Recent advances on sorting methods of high-throughput droplet-based microfluidics in enzyme directed evolution[J]. Frontiers in Chemistry, 2021, 9: 666867. |
| 16 | OBEXER R, POTT M, ZEYMER C, et al. Efficient laboratory evolution of computationally designed enzymes with low starting activities using fluorescence-activated droplet sorting[J]. Protein Engineering, Design and Selection, 2017, 30(7): 531. |
| 17 | MA F Q, CHUNG M T, YAO Y, et al. Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform[J]. Nature Communications, 2018, 9: 1030. |
| 18 | TIEMEIJER B M, DESCAMPS L, HULLEMAN J, et al. A microfluidic approach for probing heterogeneity in cytotoxic T-cells by cell pairing in hydrogel droplets[J]. Micromachines, 2022, 13(11): 1910. |
| 19 | WU L, CHEN P, DONG Y S, et al. Encapsulation of single cells on a microfluidic device integrating droplet generation with fluorescence-activated droplet sorting[J]. Biomedical Microdevices, 2013, 15(3): 553-560. |
| 20 | SUN G Y, QU L S, AZI F, et al. Recent progress in high-throughput droplet screening and sorting for bioanalysis[J]. Biosensors and Bioelectronics, 2023, 225: 115107. |
| 21 | GIELEN F, HOURS R, EMOND S, et al. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS)[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(47): E7383-E7389. |
| 22 | LINK D R, GRASLAND-MONGRAIN E, DURI A, et al. Electric control of droplets in microfluidic devices[J]. Angewandte Chemie International Edition, 2006, 45(16): 2556-2560. |
| 23 | DIEFENBACH X W, FARASAT I, GUETSCHOW E D, et al. Enabling biocatalysis by high-throughput protein engineering using droplet microfluidics coupled to mass spectrometry[J]. ACS Omega, 2018, 3(2): 1498-1508. |
| 24 | LEE K S, PALATINSZKY M, PEREIRA F C, et al. An automated Raman-based platform for the sorting of live cells by functional properties[J]. Nature Microbiology, 2019, 4(6): 1035-1048. |
| 25 | GOTO H, KANAI Y, YOTSUI A, et al. Microfluidic screening system based on boron-doped diamond electrodes and dielectrophoretic sorting for directed evolution of NAD(P)-dependent oxidoreductases[J]. Lab on a Chip, 2020, 20(4): 852-861. |
| 26 | DAVOODI H, NORDIN N, BORDONALI L, et al. An NMR-compatible microfluidic platform enabling in situ electrochemistry[J]. Lab on a Chip, 2020, 20(17): 3202-3212. |
| 27 | LABELLE C A, MASSARO A, CORTÉS-LLANOS B, et al. Image-based live cell sorting[J]. Trends in Biotechnology, 2021, 39(6): 613-623. |
| 28 | SCIAMBI A, ABATE A R. Accurate microfluidic sorting of droplets at 30 kHz[J]. Lab on a Chip, 2015, 15(1): 47-51. |
| 29 | LI S X, DING X Y, GUO F, et al. An on-chip, multichannel droplet sorter using standing surface acoustic waves[J]. Analytical Chemistry, 2013, 85(11): 5468-5474. |
| 30 | NAVI M, ABBASI N, SALARI A, et al. Magnetic water-in-water droplet microfluidics: systematic experiments and scaling mathematical analysis[J]. Biomicrofluidics, 2020, 14(2): 024101. |
| 31 | ZHONG R Y, YANG S J, STEFANO UGOLINI G, et al. Acoustofluidic droplet sorter based on single phase focused transducers (small 46/2021)[J]. Small, 2021, 17(46): e2103848. |
| 32 | ROBERT DE SAINT VINCENT M, WUNENBURGER R, DELVILLE J P. Laser switching and sorting for high speed digital microfluidics[J]. Applied Physics Letters, 2008, 92(15): 154105. |
| 33 | QIAO Y X, ZHAO X Y, ZHU J, et al. Fluorescence-activated droplet sorting of lipolytic microorganisms using a compact optical system[J]. Lab on a Chip, 2018, 18(1): 190-196. |
| 34 | TABUCHI T, YOKOBAYASHI Y. High-throughput screening of cell-free riboswitches by fluorescence-activated droplet sorting[J]. Nucleic Acids Research, 2022, 50(6): 3535-3550. |
| 35 | BECKER S. Ultra-high-throughput screening based on cell-surface display and fluorescence-activated cell sorting for the identification of novel biocatalysts[J]. Current Opinion in Biotechnology, 2004, 15(4): 323-329. |
| 36 | BASU S, CAMPBELL H M, DITTEL B N, et al. Purification of specific cell population by fluorescence activated cell sorting (FACS)[J]. Journal of Visualized Experiments: JoVE, 2010(41): 1546. |
| 37 | YUAN H L, TU R, TONG X W, et al. Ultrahigh-throughput screening of industrial enzyme-producing strains by droplet-based microfluidic system[J]. Journal of Industrial Microbiology and Biotechnology, 2022, 49(3): kuac007. |
| 38 | LARSEN A C, DUNN M R, HATCH A, et al. A general strategy for expanding polymerase function by droplet microfluidics[J]. Nature Communications, 2016, 7: 11235. |
| 39 | HASAN S, GEISSLER D, WINK K, et al. Fluorescence lifetime-activated droplet sorting in microfluidic chip systems[J]. Lab on a Chip, 2019, 19(3): 403-409. |
| 40 | HASAN S, BLAHA M E, PIENDL S K, et al. Two-photon fluorescence lifetime for label-free microfluidic droplet sorting[J].Analytical and Bioanalytical Chemistry, 2022, 414(1): 721-730. |
| 41 | HUNG S T, MUKHERJEE S, JIMENEZ R. Enrichment of rare events using a multi-parameter high throughput microfluidic droplet sorter[J]. Lab on a Chip, 2020, 20(4): 834-843. |
| 42 | NEUN S, KAMINSKI T S, HOLLFELDER F. Single-cell activity screening in microfluidic droplets[M]// Methods in enzymology: enzyme activity in single cells. Amsterdam: Elsevier, 2019: 95-112. |
| 43 | COLIN P Y, KINTSES B, GIELEN F, et al. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics[J]. Nature Communications, 2015, 6: 10008. |
| 44 | MEDCALF E J, GANTZ M, KAMINSKI T S, et al. Ultra-high-throughput absorbance-activated droplet sorting for enzyme screening at kilohertz frequencies[J]. Analytical Chemistry, 2023, 95(10): 4597-4604. |
| 45 | MACEICZYK R M, HESS D, CHIU F W Y, et al. Differential detection photothermal spectroscopy: towards ultra-fast and sensitive label-free detection in picoliter & femtoliter droplets[J]. Lab on a Chip, 2017, 17(21): 3654-3663. |
| 46 | KEMPA E E, SMITH C A, LI X, et al. Coupling droplet microfluidics with mass spectrometry for ultrahigh-throughput analysis of complex mixtures up to and above 30 Hz[J]. Analytical Chemistry, 2020, 92(18): 12605-12612. |
| 47 | WANG X X, REN L H, SU Y T, et al. Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells[J]. Analytical Chemistry, 2017, 89(22): 12569-12577. |
| 48 | WANG X X, XIN Y, REN L H, et al. Positive dielectrophoresis-based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo [J]. Science Advances, 2020, 6(32): eabb3521. |
| 49 | BEMETZ J, WEGEMANN A, SAATCHI K, et al. Microfluidic-based synthesis of magnetic nanoparticles coupled with miniaturized NMR for online relaxation studies[J]. Analytical Chemistry, 2018, 90(16): 9975-9982. |
| 50 | ANAGNOSTIDIS V, SHERLOCK B, METZ J, et al. Deep learning guided image-based droplet sorting for on-demand selection and analysis of single cells and 3D cell cultures[J]. Lab on a Chip, 2020, 20(5): 889-900. |
| 51 | RIEMER J, HOEPKEN H H, CZERWINSKA H, et al. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells[J]. Analytical Biochemistry, 2004, 331(2): 370-375. |
| 52 | HANSEN S K, JAMALI B, HUBBUCH J. Selective high throughput protein quantification based on UV absorption spectra[J]. Biotechnology and Bioengineering, 2013, 110(2): 448-460. |
| 53 | DUNCOMBE T A, PONTI A, SEEBECK F P, et al. UV-vis spectra-activated droplet sorting for label-free chemical identification and collection of droplets[J]. Analytical Chemistry, 2021, 93(38): 13008-13013. |
| 54 | HOLLAND-MORITZ D A, WISMER M K, MANN B F, et al. Mass activated droplet sorting (MADS) enables high-throughput screening of enzymatic reactions at nanoliter scale[J]. Angewandte Chemie International Edition, 2020, 59(11): 4470-4477. |
| 55 | EL-ANEED A, COHEN A, BANOUB J. Mass spectrometry, review of the basics: electrospray, MALDI, and commonly used mass analyzers[J]. Applied Spectroscopy Reviews, 2009, 44(3): 210-230. |
| 56 | HA N S, DE RAAD M, HAN L Z, et al. Faster, better, and cheaper: harnessing microfluidics and mass spectrometry for biotechnology[J]. RSC Chemical Biology, 2021, 2(5): 1331-1351. |
| 57 | SUN S W, KENNEDY R T. Droplet electrospray ionization mass spectrometry for high throughput screening for enzyme inhibitors[J]. Analytical Chemistry, 2014, 86(18): 9309-9314. |
| 58 | SUN S W, BUER B C, MARSH E N G, et al. A label-free Sirtuin 1 assay based on droplet-electrospray ionization mass spectrometry[J]. Analytical Methods, 2016, 8(17): 3458-3465. |
| 59 | STEYER D J, KENNEDY R T. High-throughput nanoelectrospray ionization-mass spectrometry analysis of microfluidic droplet samples[J]. Analytical Chemistry, 2019, 91(10): 6645-6651. |
| 60 | KUDELSKI A. Analytical applications of Raman spectroscopy[J]. Talanta, 2008, 76(1): 1-8. |
| 61 | SAFIR F, VU N, TADESSE L F, et al. Combining acoustic bioprinting with AI-assisted Raman spectroscopy for high-throughput identification of bacteria in blood[J]. Nano Letters, 2023, 23(6): 2065-2073. |
| 62 | LIU Z S, LIU D M, CAI Y D, et al. Application of nuclear magnetic resonance (NMR) in coalbed methane and shale reservoirs: a review[J]. International Journal of Coal Geology, 2020, 218: 103261. |
| 63 | CAO X Y, YANG J, MAO J D. Characterization of kerogen using solid-state nuclear magnetic resonance spectroscopy: a review[J]. International Journal of Coal Geology, 2013, 108: 83-90. |
| 64 | VAN MEERTEN S G J, VAN BENTUM P J M, KENTGENS A P M. Shim-on-chip design for microfluidic NMR detectors[J]. Analytical Chemistry, 2018, 90(17): 10134-10138. |
| 65 | SWYER I, SOONG R, DRYDEN M D M, et al. Interfacing digital microfluidics with high-field nuclear magnetic resonance spectroscopy[J]. Lab on a Chip, 2016, 16(22): 4424-4435. |
| 66 | PAN C W, HORVATH D G, BRAZA S, et al. Sorting by interfacial tension (SIFT): label-free selection of live cells based on single-cell metabolism[J]. Lab on a Chip, 2019, 19(8): 1344-1351. |
| 67 | DOBSON C, ZIELKE C, PAN C, et al. Method for passive droplet sorting after photo-tagging[J]. Micromachines, 2020, 11(11): 964. |
| 68 | ZIELKE C, PAN C W, GUTIERREZ RAMIREZ A J, et al. Microfluidic platform for the isolation of cancer-cell subpopulations based on single-cell glycolysis[J]. Analytical Chemistry, 2020, 92(10): 6949-6957. |
| 69 | WATTERSON W J, TANYERI M, WATSON A R, et al. Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes[J]. eLife, 2020, 9: e56998. |
| 70 | DOAN M, VOROBJEV I, REES P, et al. Diagnostic potential of imaging flow cytometry[J]. Trends in Biotechnology, 2018, 36(7): 649-652. |
| 71 | SUTHERLAND J D. Evolutionary optimisation of enzymes[J]. Current Opinion in Chemical Biology, 2000, 4(3): 263-269. |
| 72 | CHERRY J R, FIDANTSEF A L. Directed evolution of industrial enzymes: an update[J]. Current Opinion in Biotechnology, 2003, 14(4): 438-443. |
| 73 | TURNER N J. Directed evolution drives the next generation of biocatalysts[J]. Nature Chemical Biology, 2009, 5(8): 567-573. |
| 74 | QU G, LI A T, ACEVEDO-ROCHA C G, et al. The crucial role of methodology development in directed evolution of selective enzymes[J]. Angewandte Chemie International Edition, 2020, 59(32): 13204-13231. |
| 75 | OTTEN R, PÁDUA R A P, BUNZEL H A, et al. How directed evolution reshapes the energy landscape in an enzyme to boost catalysis[J]. Science, 2020, 370(6523): 1442-1446. |
| 76 | WANG Y J, XUE P, CAO M F, et al. Directed evolution: methodologies and applications[J]. Chemical Reviews, 2021, 121(20): 12384-12444. |
| 77 | PACKER M S, LIU D R. Methods for the directed evolution of proteins[J]. Nature Reviews Genetics, 2015, 16(7): 379-394. |
| 78 | CHIU F W Y, STAVRAKIS S. High-throughput droplet-based microfluidics for directed evolution of enzymes[J]. ELECTROPHORESIS, 2019, 40(21): 2860-2872. |
| 79 | MADHAVAN A, ARUN K B, BINOD P, et al. Design of novel enzyme biocatalysts for industrial bioprocess: harnessing the power of protein engineering, high throughput screening and synthetic biology[J]. Bioresource Technology, 2021, 325: 124617. |
| 80 | VAN LOO B, HEBERLEIN M, MAIR P, et al. High-throughput, lysis-free screening for sulfatase activity using Escherichia coli autodisplay in microdroplets[J]. ACS Synthetic Biology, 2019, 8(12): 2690-2700. |
| 81 | HE R L, DING R H, HEYMAN J A, et al. Ultra-high-throughput picoliter-droplet microfluidics screening of the industrial cellulase-producing filamentous fungus Trichoderma reesei [J]. Journal of Industrial Microbiology and Biotechnology, 2019, 46(11): 1603-1610. |
| 82 | PRODANOVIĆ R, UNG W L, ILIĆ ĐURĐIĆ K, et al. A high-throughput screening system based on droplet microfluidics for glucose oxidase gene libraries[J]. Molecules, 2020, 25(10): 2418. |
| 83 | ZUREK P J, KNYPHAUSEN P, NEUFELD K, et al. UMI-linked consensus sequencing enables phylogenetic analysis of directed evolution[J]. Nature Communications, 2020, 11: 6023. |
| 84 | SUN G Y, WU Y K, HUANG Z Y, et al. Directed evolution of diacetylchitobiose deacetylase via high-throughput droplet sorting with a novel, bacteria-based biosensor[J]. Biosensors and Bioelectronics, 2023, 219: 114818. |
| 85 | QIAO Y, HU R, CHEN D, et al. Fluorescence-activated droplet sorting of PET degrading microorganisms[J]. Journal of Hazardous Materials, 2022, 424(pt b): 127417. |
| 86 | XU A M, ZHANG X X, CAO S X, et al. Transcription-associated fluorescence-activated droplet sorting for di-rhamnolipid hyperproducers[J]. ACS Synthetic Biology, 2022, 11(6): 1992-2000. |
| 87 | ZACHOS I, GENTH R, SUTIONO S, et al. Hot flows: evolving an archaeal glucose dehydrogenase for ultrastable carba-NADP+ using microfluidics at elevated temperatures[J]. ACS Catalysis, 2022, 12(3): 1841-1846. |
| 88 | TAN Y M, ZHANG Y, HAN Y B, et al. Directed evolution of an α1,3-fucosyltransferase using a single-cell ultrahigh-throughput screening method[J]. Science Advances, 2019, 5(10): eaaw8451. |
| 89 | KAMINSKI T S, SCHELER O, GARSTECKI P. Droplet microfluidics for microbiology: techniques, applications and challenges[J]. Lab on a Chip, 2016, 16(12): 2168-2187. |
| 90 | BOWMAN E K, NGUYEN HOANG P T, GORDILLO SIERRA A R, et al. Temporal sorting of microdroplets can identify productivity differences of itaconic acid from libraries of Yarrowia lipolytica [J]. Lab on a Chip, 2023, 23(9): 2249-2256. |
| 91 | AN X S, ZUO P, YE B C. A single cell droplet microfluidic system for quantitative determination of food-borne pathogens[J]. Talanta, 2020, 209: 120571. |
| 92 | NEETHIRAJAN S, KOBAYASHI I, NAKAJIMA M, et al. Microfluidics for food, agriculture and biosystems industries[J]. Lab on a Chip, 2011, 11(9): 1574-1586. |
| 93 | XING G W, ZHANG W F, LI N, et al. Recent progress on microfluidic biosensors for rapid detection of pathogenic bacteria[J]. Chinese Chemical Letters, 2022, 33(4): 1743-1751. |
| 94 | LI S X, MA F, BACHMAN H, et al. Acoustofluidic bacteria separation[J]. Journal of Micromechanics and Microengineering, 2017, 27(1): 015031. |
| 95 | OHLSSON P, PETERSSON K, AUGUSTSSON P, et al. Acoustic impedance matched buffers enable separation of bacteria from blood cells at high cell concentrations[J]. Scientific Reports, 2018, 8: 9156. |
| 96 | HYMAN L B, CHRISTOPHER C R, ROMERO P A. Single-cell nucleic acid profiling in droplets (SNAPD) enables high-throughput analysis of heterogeneous cell populations[J]. Nucleic Acids Research, 2021, 49(18): e103. |
| 97 | SEAH Y F S, HU H X, MERTEN C A. Microfluidic single-cell technology in immunology and antibody screening[J]. Molecular Aspects of Medicine, 2018, 59: 47-61. |
| 98 | MAZUTIS L, GILBERT J, UNG W L, et al. Single-cell analysis and sorting using droplet-based microfluidics[J]. Nature Protocols, 2013, 8(5): 870-891. |
| 99 | BARANOVA M N, BABIKOVA P A, KUDZHAEV A M, et al. Live biosensors for ultrahigh-throughput screening of antimicrobial activity against gram-negative bacteria[J]. Antibiotics, 2021, 10(10): 1161. |
| 100 | SILTANEN C A, COLE R H, POUST S, et al. An oil-free picodrop bioassay platform for synthetic biology[J]. Scientific Reports, 2018, 8: 7913. |
| [1] | GUO Xiaojie, JIAN Xingjin, WANG Liyan, ZHANG Chong, XING Xinhui. Progress in bioreactors and instruments for phenotype testing with synthetic biology research [J]. Synthetic Biology Journal, 2024, 5(1): 16-37. |
| [2] | Huan LIU, Qiu CUI. Advances and applications of ambient ionization mass spectrometry in screening of microbial strains [J]. Synthetic Biology Journal, 2023, 4(5): 980-999. |
| [3] | WU Yujie, LIU Xinxin, LIU Jianhui, Yang Kaiguang, SUI Zhigang, ZHANG Lihua, ZHANG Yukui. Research progress of strain screening and quantitative analysis of key molecules based on high-throughput liquid chromatography and mass spectrometry [J]. Synthetic Biology Journal, 2023, 4(5): 1000-1019. |
| [4] | ZHAO Guomiao, YANG Xin, ZHANG Yuan, WANG Jing, TAN Jian, WEI Chao, ZHOU Nana, LI Fan, WANG Xiaoyan. Biofoundry and its industrial application [J]. Synthetic Biology Journal, 2023, 4(5): 892-903. |
| [5] | Ran TU, Shixin LI, Haoni LI, Meng WANG. Advances and applications of droplet-based microfluidics in evolution and screening of engineered microbial strains [J]. Synthetic Biology Journal, 2023, 4(1): 165-184. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||


