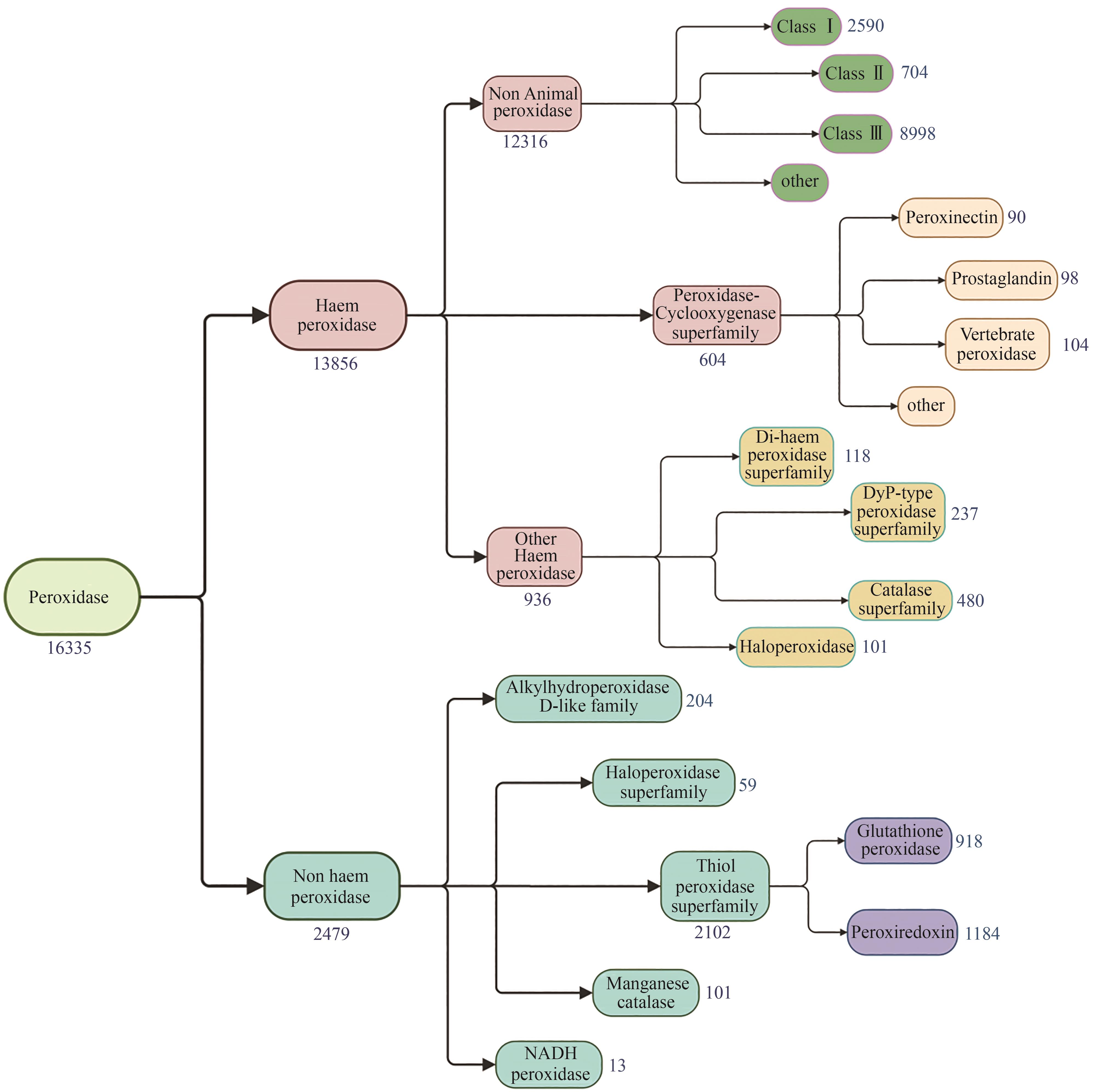
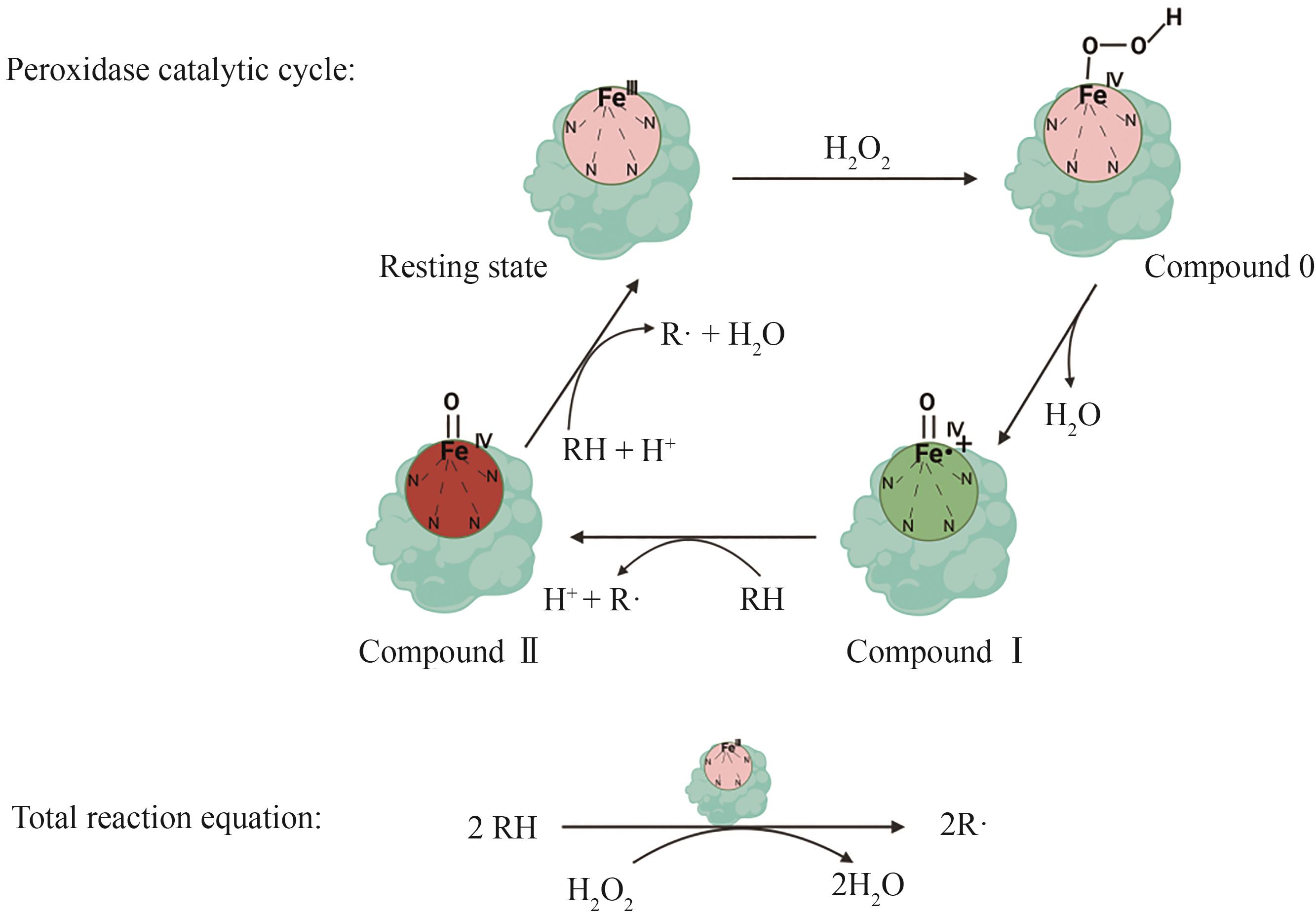
Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (6): 1498-1517.DOI: 10.12211/2096-8280.2024-027
• Invited Review • Previous Articles Next Articles
Research progress in recombinant expression and application of peroxidases
LI Geng, SHEN Xiaolin, SUN Xinxiao, WANG Jia, YUAN Qipeng
- State Key Laboratory of Chemical Resource Engineering,College of Life Science and Biotechnology,Beijing University of Chemical Technology,Beijing 100029,China
-
Received:2024-03-21Revised:2024-06-28Online:2025-01-10Published:2024-12-31 -
Contact:WANG Jia, YUAN Qipeng
过氧化物酶的重组表达和应用研究进展
李庚, 申晓林, 孙新晓, 王佳, 袁其朋
- 北京化工大学生命科学与技术学院,化工资源有效利用国家重点实验室,北京 100029
-
通讯作者:王佳,袁其朋 -
作者简介:李庚 (1998—),男,硕士研究生。研究方向为代谢工程及合成生物学。E-mail:1602975489@qq.com王佳 (1989—),女,博士,教授。研究方向为代谢工程及合成生物学。E-mail:wangjia@mail.buct.edu.cn袁其朋 (1969—),男,博士,教授。研究方向为生物化工,代谢工程及微生物合成生物学。E-mail:yuanqp@mail.buct.edu.cn -
基金资助:国家重点研发计划(2022YFC2106100);国家自然科学基金(22378016)
CLC Number:
Cite this article
LI Geng, SHEN Xiaolin, SUN Xinxiao, WANG Jia, YUAN Qipeng. Research progress in recombinant expression and application of peroxidases[J]. Synthetic Biology Journal, 2024, 5(6): 1498-1517.
李庚, 申晓林, 孙新晓, 王佳, 袁其朋. 过氧化物酶的重组表达和应用研究进展[J]. 合成生物学, 2024, 5(6): 1498-1517.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-027
| 表达宿主 | 过氧化物酶 | 表达方式和策略 | 表达效果 | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌 | HRP | 周质表达,通过定向进化提高酶表达活力 | 表达量110 mg/L 酶活力140 U/L | [ |
| 大肠杆菌 | HRP | 胞内表达,纯化的包涵体通过体外重折叠复性 | 比酶活10 U/mg | [ |
| 大肠杆菌 | HRP | 胞内表达,纯化的包涵体通过体外重折叠复性 | 表达量1000 mg/L 比酶活62.5 U/mg | [ |
| 大肠杆菌 | HRP | 周质表达,通过将HRP的N末端与DsbA蛋白融合,利用SRP途径将融合蛋白转运至周质 | 表达量48 mg/L 比酶活12.7 U/mg | [ |
| 大肠杆菌 | HRP | 胞内表达,通过与甘油酸激酶(PGK)融合表达实现可溶性过量表达,重组蛋白与钙离子、氯化血红素和氧化型谷胱甘肽一起简单孵育后激活活性 | 酶回收量72 mg/L 比酶活为商业酶的60% | [ |
| 大肠杆菌 | MnP | 胞内表达,纯化的包涵体通过体外重折叠复性 | 酶活力345 U/L 比酶活3.63 U/mg | [ |
| 大肠杆菌 | MnP | 胞内表达,通过在细胞培养物中加入化学物质(0.25% Triton X-100、0.25% Tween-80、0.5%甘油和1%乙醇)改善蛋白溶解度 | 酶活力310 U/L 比酶活0.441 U/mg | [ |
| 大肠杆菌 | MnP | 胞内表达,通过密码子优化、冷休克启动子的控制表达、培养过程中连续添加血红素及共表达伴侣蛋白改善蛋白溶解度 | 表达量30 mg/L kcat/Km=406 L/(s·mmol) | [ |
| 大肠杆菌 | MnP | 胞内表达,通过共表达二硫键异构酶DsbC改善MnP的溶解度 | 比酶活445 U/mg | [ |
| 毕赤酵母 | MnP | 周质表达,与α因子前导序列的融合表达 | 酶活力48 U/L 比酶活42 U/mg | [ |
| 酿酒酵母 | HRP | 周质表达,通过定向进化提高酶表达活力 | 酶活力1080 U/L | [ |
| 毕赤酵母 | HRP | 周质表达,通过定向进化提高酶表达活力 | 表达量6 mg/L 比酶活980 U/mg | [ |
| 毕赤酵母 | HRP | 周质表达,通过敲除α-1,6-甘露糖基转移酶(OCH1编码)改善HRP高糖基化问题,通过发酵优化提高表达水平 | 表达量101 mg/L 比酶活278.1 U/mg | [ |
| 毕赤酵母 | HRP | 周质表达,通过与链球菌蛋白G(SpG)融合表达提高表达量和比酶活 | 表达量113 mg/L 比酶活1624 U/mg | [ |
| 毕赤酵母 | HRP | 周质表达,OCH1缺陷的毕赤酵母菌株中表达了通过定向进化得到的突变体HRP 13A7-N175S | 表达量132 mg/L 比酶活2008 U/mg | [ |
| 黑曲霉 | MnP | 周质表达,共表达钙连蛋白,培养基添加血红素 | 表达量70 mg/L | [ |
| 米曲霉 | MnP | 周质表达,在Taka淀粉酶启动子下表达融合了分泌信号的MnP | 表达效率0.33 U/(min·L) kcat=132 s-1 | [ |
Table 1 Research progress of recombinant peroxidase expression
| 表达宿主 | 过氧化物酶 | 表达方式和策略 | 表达效果 | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌 | HRP | 周质表达,通过定向进化提高酶表达活力 | 表达量110 mg/L 酶活力140 U/L | [ |
| 大肠杆菌 | HRP | 胞内表达,纯化的包涵体通过体外重折叠复性 | 比酶活10 U/mg | [ |
| 大肠杆菌 | HRP | 胞内表达,纯化的包涵体通过体外重折叠复性 | 表达量1000 mg/L 比酶活62.5 U/mg | [ |
| 大肠杆菌 | HRP | 周质表达,通过将HRP的N末端与DsbA蛋白融合,利用SRP途径将融合蛋白转运至周质 | 表达量48 mg/L 比酶活12.7 U/mg | [ |
| 大肠杆菌 | HRP | 胞内表达,通过与甘油酸激酶(PGK)融合表达实现可溶性过量表达,重组蛋白与钙离子、氯化血红素和氧化型谷胱甘肽一起简单孵育后激活活性 | 酶回收量72 mg/L 比酶活为商业酶的60% | [ |
| 大肠杆菌 | MnP | 胞内表达,纯化的包涵体通过体外重折叠复性 | 酶活力345 U/L 比酶活3.63 U/mg | [ |
| 大肠杆菌 | MnP | 胞内表达,通过在细胞培养物中加入化学物质(0.25% Triton X-100、0.25% Tween-80、0.5%甘油和1%乙醇)改善蛋白溶解度 | 酶活力310 U/L 比酶活0.441 U/mg | [ |
| 大肠杆菌 | MnP | 胞内表达,通过密码子优化、冷休克启动子的控制表达、培养过程中连续添加血红素及共表达伴侣蛋白改善蛋白溶解度 | 表达量30 mg/L kcat/Km=406 L/(s·mmol) | [ |
| 大肠杆菌 | MnP | 胞内表达,通过共表达二硫键异构酶DsbC改善MnP的溶解度 | 比酶活445 U/mg | [ |
| 毕赤酵母 | MnP | 周质表达,与α因子前导序列的融合表达 | 酶活力48 U/L 比酶活42 U/mg | [ |
| 酿酒酵母 | HRP | 周质表达,通过定向进化提高酶表达活力 | 酶活力1080 U/L | [ |
| 毕赤酵母 | HRP | 周质表达,通过定向进化提高酶表达活力 | 表达量6 mg/L 比酶活980 U/mg | [ |
| 毕赤酵母 | HRP | 周质表达,通过敲除α-1,6-甘露糖基转移酶(OCH1编码)改善HRP高糖基化问题,通过发酵优化提高表达水平 | 表达量101 mg/L 比酶活278.1 U/mg | [ |
| 毕赤酵母 | HRP | 周质表达,通过与链球菌蛋白G(SpG)融合表达提高表达量和比酶活 | 表达量113 mg/L 比酶活1624 U/mg | [ |
| 毕赤酵母 | HRP | 周质表达,OCH1缺陷的毕赤酵母菌株中表达了通过定向进化得到的突变体HRP 13A7-N175S | 表达量132 mg/L 比酶活2008 U/mg | [ |
| 黑曲霉 | MnP | 周质表达,共表达钙连蛋白,培养基添加血红素 | 表达量70 mg/L | [ |
| 米曲霉 | MnP | 周质表达,在Taka淀粉酶启动子下表达融合了分泌信号的MnP | 表达效率0.33 U/(min·L) kcat=132 s-1 | [ |
| 污染物类型 | 降解底物 | 过氧化物酶 | 研究成果 | 参考文献 |
|---|---|---|---|---|
| 酚类污染物 | 苯酚 | 芜菁过氧化物酶(TP) | 采用海藻酸盐包埋分离于芜菁根的TP,在最佳条件下对苯酚的平均脱除率达93% | [ |
| 苯酚 | 辣根过氧化物酶(HRP) | 采用一步包埋法将HRP酶固定在半透性海藻酸盐膜,在重复使用5次时酶活下降至50% | [ | |
| 苯酚 | 辣根过氧化物酶(HRP) | 在改性丙烯腈共聚物膜上共价固定HRP,对100 mg/L的苯酚溶液脱除率达95.4%,具备高稳定性,在酶操作的20天后酶活下降至50% | [ | |
| 苯酚、对甲酚 | 黑萝卜过氧化物酶(RSVNP) | 以交联酶聚集体形式固定化的RSVNP-CLEA具备高稳定性,储存60天后保持100%的活性,最佳条件下显示出对苯酚和对甲酚92%和98%的高消除率 | [ | |
| 苯酚、对氯苯酚 | 辣根过氧化物酶(HRP) | HRP通过戊二醛偶联共价固定在磁珠上,固定化HRP保留了79%游离酶活性,在磁稳定流化床反应器中能够消除100%的苯酚和92%对氯苯酚 | [ | |
| 苯酚 | 辣根过氧化物酶(HRP) | 物理固定在改性还原氧化石墨烯纳米颗粒(NP)的HRP显示出对高浓度苯酚(2.5 g/L)100%的去除率,重复使用10次后仍能保留60%的初始酶活性 | [ | |
| 对溴苯酚 | 苦瓜过氧化物酶(BGP) | Con A层状藻酸钙纤维素珠为载体固定的BGP在填充床反应器中能够清除96%的对溴苯酚,而且连续运行30天后仍能保持75%的清除率 | [ | |
| 苯酚 | 辣根过氧化物酶(HRP) | 通过戊二醛交联剂固定在生物碳上的HRP具备高稳定性,重复使用10次后仍保持70%的酶活性 | [ | |
| 苯酚、对氯苯酚 | 辣根过氧化物酶(HRP) | 固定在阳离子微孔淀粉的HRP显示出较游离酶更强的苯酚和对氯苯酚的去除能力,重复使用10次后仍保留66%的初始活性 | [ | |
| 苯酚 | 辣根过氧化物酶(HRP) | 以氧化海藻酸酪胺微珠为载体固定的HRP在最佳条件下能够去除96%的苯酚,重复使用4次后仍保留61%的初始活性 | [ | |
| 合成染料 | 甲基橙 | 辣根过氧化物酶(HRP) | 采用聚丙烯酰胺凝胶包埋提取于新鲜辣根的HRP,在填充床生物反应器中能够降解90%的甲基橙 | [ |
| 酚红 | 辣根过氧化物酶(HRP) | 采用了包埋和共价键合组合方法将HRP固定在海藻酸钙-淀粉杂化小球上,固定化HRP对酚红的去除率为55.87%,是游离HRP的11.5倍 | [ | |
| 黑色CKF、蓝色GWF、红色C4BLN | 来自裂褶菌IBL-06的木质素过氧化物酶(LiP) | 以戊二醛为交联剂将LiP固定化得到的交联酶聚集体能够有效降解黑色CKF、蓝色GWF和红色C4BLN三种活性染料,处理6 h后的降解率分别为89.6%、81.46%和79.6% | [ | |
| 晶状丽春红6R(CP6R) | 大豆过氧化物酶(SBP) | 40 mg/L的CP6R染料溶液可以在1 min内被SBP完全降解,进一步研究结果表明偶氮染料通过对称和不对称偶氮键断裂两种途径降解 | [ | |
| 结晶紫 | 来源于解淀粉芽孢杆菌MN-13的过氧化物酶(BaDyP) | BaDyP处理结晶紫(50 mg/L)72 h后,降解率为63.0% | [ | |
| 偶氮料DY106 | 西葫芦过氧化物酶(CP) | 最佳条件下,海藻酸钙包埋的CP能够在2 min内使87%以上的染料DY106(50 mg/L)脱色 | [ | |
| 刚果红、甲基橙、铬黑T、 | 来源于白腐真菌Phanerochaete sp. HSD的锰过氧化物酶(MnP) | 菌液中提取的MnP粗酶能有效降解刚果红、甲基橙和铬黑三种偶氮染料,在数小时内能够将浓度200 mg/L以上的染料降解90%以上 | [ | |
| 台盼蓝 | 大豆过氧化物酶(SBP) | 聚丙烯酰胺(PA)包埋的SBP具备更好的稳定性和可重复使用性,在30 min内能够使台盼蓝(40 mg/L)降解约80% | [ | |
| 偶氮染料直红23和直蓝15 | 来源于天蓝色链霉菌SPR7的过氧化物酶POX | 在添加10 mmol/L木质素的条件下,POX经过20 h能够使偶氮染料直红23和直蓝15脱色75.4%和90% | [ | |
| 酸性蓝158(AB) | 黑曲霉CTM10002来源的锰过氧化物酶MnP AN30 | 在反应24 h后,纯化的MnP AN30对染料AB的脱色率达到98% | [ | |
| 绿松石蓝133G、Drim Red CL4BN | 花椰菜过氧化物酶(CFP) | 提取自新鲜花椰菜叶的CFP在最佳条件下能够使染料绿松石蓝133G(25 mg/L)和Drim Red CL4BN(50 mg/L)降解85%和92% | [ | |
| 药物活性化合物 | 17β-雌激素(E2) | 辣根过氧化物酶(HRP) | 以磁性Fe3O4为载体固定的HRP在最佳条件下能够使E2(1000 mg/L)降解80% | [ |
| 双酚a(BPA)、三氯生(TCS)、雌酮(E1)、17β雌二醇(E2)和17α炔雌醇(EE2) | 黑管菌来源的多功能过氧化物酶(VP) | 分离于黑管菌的VP能够高效去除5种污染物,去除速率达到[2.5~5.0 mg/(L·min)] | [ | |
| 三氯生(TCS)、磺胺甲𫫇唑(SMX)、雌酮(E1)、17β-雌二醇(E2)和17α-炔雌醇(EE2) | 大豆过氧化物酶(SBP) | 在优化的条件下,SBP对浓度各10 μmol/L的药物污染物废水降解率在95%以上 | [ | |
| 磺胺甲𫫇唑(SMX) | 大豆过氧化物酶(SBP) | 在78 nmol/LSBP和56 μmol/L H2O2的条件下反应3 h,浓度为500 mg/L的SMX能够降解78% | [ | |
| 双氯芬酸、卡马西平和扑热息痛 | 辣根过氧化物酶(HRP)和木质素过氧化物酶(LiP) | 溶胶-凝胶包封的HRP和LiP酶复合物都能够改善酶在酸性介质中的稳定性,而且在反应3天后可以完全降解双氯芬酸、卡马西平和扑热息痛 | [ | |
| 四环素(TC)和土霉素(OTC) | 来源于黄孢原毛平革菌的过氧化物酶(MnP) | 添加40 U/L的MnP时,50 mg/L的TC和OTC能够在4 h内降解72.5%和84.3% | [ | |
| 磺胺甲𫫇唑(SMX)和卡马西平(CBZ) | 辣根过氧化物酶(HRP) | 由聚氯乙烯电纺丝纤维封装固定化的HRP显著提升了其稳定性和重复使用性,而且能够在24 h内降解80%以上10 mg/L的SMZ和CBZ | [ | |
| 磺胺甲𫫇唑、萘普生、四环素、雌二醇、酮康唑、酮咯酸和双氯芬酸 | 来源于烟曲霉的氯过氧化物酶(CPO) | CPO能够氧化磺胺甲𫫇唑、萘普生、四环素、雌二醇、酮康唑、酮咯酸和双氯芬酸,反应10 min后,所有底物均能转化80%以上 | [ |
Table 2 Studies on the degradation of organic pollutants by peroxidases
| 污染物类型 | 降解底物 | 过氧化物酶 | 研究成果 | 参考文献 |
|---|---|---|---|---|
| 酚类污染物 | 苯酚 | 芜菁过氧化物酶(TP) | 采用海藻酸盐包埋分离于芜菁根的TP,在最佳条件下对苯酚的平均脱除率达93% | [ |
| 苯酚 | 辣根过氧化物酶(HRP) | 采用一步包埋法将HRP酶固定在半透性海藻酸盐膜,在重复使用5次时酶活下降至50% | [ | |
| 苯酚 | 辣根过氧化物酶(HRP) | 在改性丙烯腈共聚物膜上共价固定HRP,对100 mg/L的苯酚溶液脱除率达95.4%,具备高稳定性,在酶操作的20天后酶活下降至50% | [ | |
| 苯酚、对甲酚 | 黑萝卜过氧化物酶(RSVNP) | 以交联酶聚集体形式固定化的RSVNP-CLEA具备高稳定性,储存60天后保持100%的活性,最佳条件下显示出对苯酚和对甲酚92%和98%的高消除率 | [ | |
| 苯酚、对氯苯酚 | 辣根过氧化物酶(HRP) | HRP通过戊二醛偶联共价固定在磁珠上,固定化HRP保留了79%游离酶活性,在磁稳定流化床反应器中能够消除100%的苯酚和92%对氯苯酚 | [ | |
| 苯酚 | 辣根过氧化物酶(HRP) | 物理固定在改性还原氧化石墨烯纳米颗粒(NP)的HRP显示出对高浓度苯酚(2.5 g/L)100%的去除率,重复使用10次后仍能保留60%的初始酶活性 | [ | |
| 对溴苯酚 | 苦瓜过氧化物酶(BGP) | Con A层状藻酸钙纤维素珠为载体固定的BGP在填充床反应器中能够清除96%的对溴苯酚,而且连续运行30天后仍能保持75%的清除率 | [ | |
| 苯酚 | 辣根过氧化物酶(HRP) | 通过戊二醛交联剂固定在生物碳上的HRP具备高稳定性,重复使用10次后仍保持70%的酶活性 | [ | |
| 苯酚、对氯苯酚 | 辣根过氧化物酶(HRP) | 固定在阳离子微孔淀粉的HRP显示出较游离酶更强的苯酚和对氯苯酚的去除能力,重复使用10次后仍保留66%的初始活性 | [ | |
| 苯酚 | 辣根过氧化物酶(HRP) | 以氧化海藻酸酪胺微珠为载体固定的HRP在最佳条件下能够去除96%的苯酚,重复使用4次后仍保留61%的初始活性 | [ | |
| 合成染料 | 甲基橙 | 辣根过氧化物酶(HRP) | 采用聚丙烯酰胺凝胶包埋提取于新鲜辣根的HRP,在填充床生物反应器中能够降解90%的甲基橙 | [ |
| 酚红 | 辣根过氧化物酶(HRP) | 采用了包埋和共价键合组合方法将HRP固定在海藻酸钙-淀粉杂化小球上,固定化HRP对酚红的去除率为55.87%,是游离HRP的11.5倍 | [ | |
| 黑色CKF、蓝色GWF、红色C4BLN | 来自裂褶菌IBL-06的木质素过氧化物酶(LiP) | 以戊二醛为交联剂将LiP固定化得到的交联酶聚集体能够有效降解黑色CKF、蓝色GWF和红色C4BLN三种活性染料,处理6 h后的降解率分别为89.6%、81.46%和79.6% | [ | |
| 晶状丽春红6R(CP6R) | 大豆过氧化物酶(SBP) | 40 mg/L的CP6R染料溶液可以在1 min内被SBP完全降解,进一步研究结果表明偶氮染料通过对称和不对称偶氮键断裂两种途径降解 | [ | |
| 结晶紫 | 来源于解淀粉芽孢杆菌MN-13的过氧化物酶(BaDyP) | BaDyP处理结晶紫(50 mg/L)72 h后,降解率为63.0% | [ | |
| 偶氮料DY106 | 西葫芦过氧化物酶(CP) | 最佳条件下,海藻酸钙包埋的CP能够在2 min内使87%以上的染料DY106(50 mg/L)脱色 | [ | |
| 刚果红、甲基橙、铬黑T、 | 来源于白腐真菌Phanerochaete sp. HSD的锰过氧化物酶(MnP) | 菌液中提取的MnP粗酶能有效降解刚果红、甲基橙和铬黑三种偶氮染料,在数小时内能够将浓度200 mg/L以上的染料降解90%以上 | [ | |
| 台盼蓝 | 大豆过氧化物酶(SBP) | 聚丙烯酰胺(PA)包埋的SBP具备更好的稳定性和可重复使用性,在30 min内能够使台盼蓝(40 mg/L)降解约80% | [ | |
| 偶氮染料直红23和直蓝15 | 来源于天蓝色链霉菌SPR7的过氧化物酶POX | 在添加10 mmol/L木质素的条件下,POX经过20 h能够使偶氮染料直红23和直蓝15脱色75.4%和90% | [ | |
| 酸性蓝158(AB) | 黑曲霉CTM10002来源的锰过氧化物酶MnP AN30 | 在反应24 h后,纯化的MnP AN30对染料AB的脱色率达到98% | [ | |
| 绿松石蓝133G、Drim Red CL4BN | 花椰菜过氧化物酶(CFP) | 提取自新鲜花椰菜叶的CFP在最佳条件下能够使染料绿松石蓝133G(25 mg/L)和Drim Red CL4BN(50 mg/L)降解85%和92% | [ | |
| 药物活性化合物 | 17β-雌激素(E2) | 辣根过氧化物酶(HRP) | 以磁性Fe3O4为载体固定的HRP在最佳条件下能够使E2(1000 mg/L)降解80% | [ |
| 双酚a(BPA)、三氯生(TCS)、雌酮(E1)、17β雌二醇(E2)和17α炔雌醇(EE2) | 黑管菌来源的多功能过氧化物酶(VP) | 分离于黑管菌的VP能够高效去除5种污染物,去除速率达到[2.5~5.0 mg/(L·min)] | [ | |
| 三氯生(TCS)、磺胺甲𫫇唑(SMX)、雌酮(E1)、17β-雌二醇(E2)和17α-炔雌醇(EE2) | 大豆过氧化物酶(SBP) | 在优化的条件下,SBP对浓度各10 μmol/L的药物污染物废水降解率在95%以上 | [ | |
| 磺胺甲𫫇唑(SMX) | 大豆过氧化物酶(SBP) | 在78 nmol/LSBP和56 μmol/L H2O2的条件下反应3 h,浓度为500 mg/L的SMX能够降解78% | [ | |
| 双氯芬酸、卡马西平和扑热息痛 | 辣根过氧化物酶(HRP)和木质素过氧化物酶(LiP) | 溶胶-凝胶包封的HRP和LiP酶复合物都能够改善酶在酸性介质中的稳定性,而且在反应3天后可以完全降解双氯芬酸、卡马西平和扑热息痛 | [ | |
| 四环素(TC)和土霉素(OTC) | 来源于黄孢原毛平革菌的过氧化物酶(MnP) | 添加40 U/L的MnP时,50 mg/L的TC和OTC能够在4 h内降解72.5%和84.3% | [ | |
| 磺胺甲𫫇唑(SMX)和卡马西平(CBZ) | 辣根过氧化物酶(HRP) | 由聚氯乙烯电纺丝纤维封装固定化的HRP显著提升了其稳定性和重复使用性,而且能够在24 h内降解80%以上10 mg/L的SMZ和CBZ | [ | |
| 磺胺甲𫫇唑、萘普生、四环素、雌二醇、酮康唑、酮咯酸和双氯芬酸 | 来源于烟曲霉的氯过氧化物酶(CPO) | CPO能够氧化磺胺甲𫫇唑、萘普生、四环素、雌二醇、酮康唑、酮咯酸和双氯芬酸,反应10 min后,所有底物均能转化80%以上 | [ |
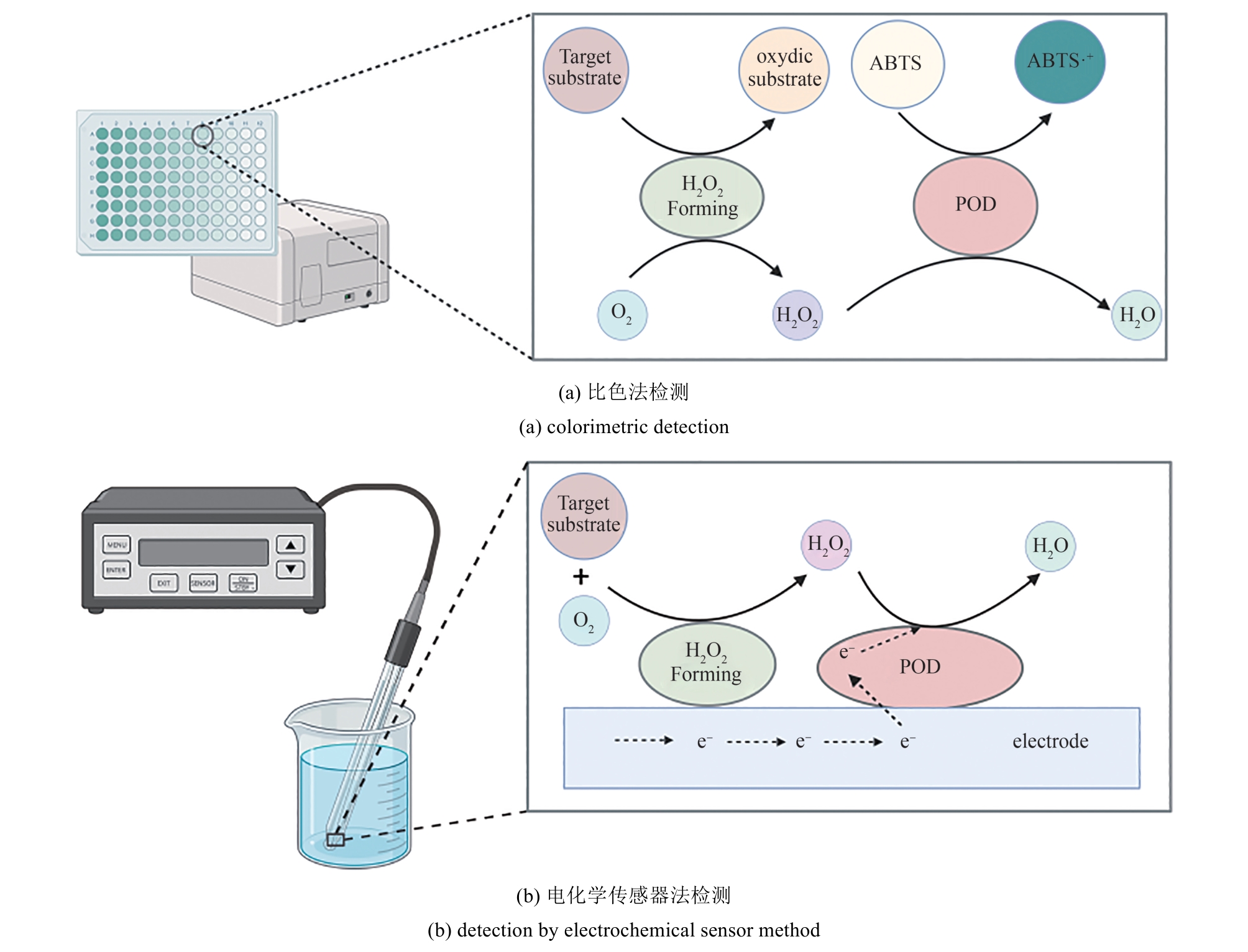
| 检测目标物 | H2O2生成酶 | 检测原理 | 参考文献 |
|---|---|---|---|
| 乳糖 | β-半乳糖苷酶+半乳糖氧化酶 | 表面共固定β-半乳糖苷酶、半乳糖氧化酶和POD的石墨电极电流传感器 | [ |
| 乳糖 | β-半乳糖苷酶+葡萄糖氧化酶 | POD催化ABTS显色 | [ |
| 尿酸 | 尿酸氧化酶 | POD催化TMB显色 | [ |
| 尿酸 | 尿酸氧化酶 | 表面固定HRP的聚四氟乙烯膜电流传感器 | [ |
| 葡萄糖 | 葡萄糖氧化酶 | HRP刻蚀金纳米棒(AuNR)显色 | [ |
| 葡萄糖 | 葡萄糖氧化酶 | 表面固定HRP的碳纤维电流传感器 | [ |
| 甘油三酯 | 脂肪酶+甘油激酶+3-磷酸甘油氧化酶 | HRP催化3,5-二氯-2-羟基苯甲酸和4-氨基苯乙酮聚合显色 | [ |
| 甘油三酯 | 脂肪酶+甘油激酶+3-磷酸甘油氧化酶 | 表面固定功能化酶聚集体的多晶金电极电流传感器 | [ |
| 胆固醇 | 胆固醇氧化酶 | HRP催化ABTS显色 | [ |
| 胆固醇 | 胆固醇氧化酶+胆固醇酯酶 | 表面共固定胆固醇氧化酶、胆固醇酯酶和POD的聚苯胺膜电流传感器 | [ |
| 酚类化合物 | H2O2直接添加 | 表面固定POD的石墨电极电流传感器 | [ |
Table 3 POD-based methods for the detection of compounds
| 检测目标物 | H2O2生成酶 | 检测原理 | 参考文献 |
|---|---|---|---|
| 乳糖 | β-半乳糖苷酶+半乳糖氧化酶 | 表面共固定β-半乳糖苷酶、半乳糖氧化酶和POD的石墨电极电流传感器 | [ |
| 乳糖 | β-半乳糖苷酶+葡萄糖氧化酶 | POD催化ABTS显色 | [ |
| 尿酸 | 尿酸氧化酶 | POD催化TMB显色 | [ |
| 尿酸 | 尿酸氧化酶 | 表面固定HRP的聚四氟乙烯膜电流传感器 | [ |
| 葡萄糖 | 葡萄糖氧化酶 | HRP刻蚀金纳米棒(AuNR)显色 | [ |
| 葡萄糖 | 葡萄糖氧化酶 | 表面固定HRP的碳纤维电流传感器 | [ |
| 甘油三酯 | 脂肪酶+甘油激酶+3-磷酸甘油氧化酶 | HRP催化3,5-二氯-2-羟基苯甲酸和4-氨基苯乙酮聚合显色 | [ |
| 甘油三酯 | 脂肪酶+甘油激酶+3-磷酸甘油氧化酶 | 表面固定功能化酶聚集体的多晶金电极电流传感器 | [ |
| 胆固醇 | 胆固醇氧化酶 | HRP催化ABTS显色 | [ |
| 胆固醇 | 胆固醇氧化酶+胆固醇酯酶 | 表面共固定胆固醇氧化酶、胆固醇酯酶和POD的聚苯胺膜电流传感器 | [ |
| 酚类化合物 | H2O2直接添加 | 表面固定POD的石墨电极电流传感器 | [ |
| 1 | SAVELLI B, LI Q, WEBBER M, et al. RedoxiBase: a database for ROS homeostasis regulated proteins[J]. Redox Biology, 2019, 26: 101247. |
| 2 | ZAMOCKY M, JAKOPITSCH C, FURTMÜLLER P G, et al. The peroxidase-cyclooxygenase superfamily: reconstructed evolution of critical enzymes of the innate immune system[J]. Proteins: Structure, Function, and Bioinformatics, 2008, 72(2): 589-605. |
| 3 | KANGASJÄRVI S, LEPISTÖ A, HÄNNIKÄINEN K, et al. Diverse roles for chloroplast stromal and thylakoid-bound ascorbate peroxidases in plant stress responses[J]. The Biochemical Journal, 2008, 412(2): 275-285. |
| 4 | PREETHI P S, PATHY M R. Recombinant peroxidase: production and its prospective applications-a review[J]. Research Journal of Pharmacy and Technology, 2018, 11(7): 3186-3196. |
| 5 | MARTINS D, KATHIRESAN M, ENGLISH A M. Cytochrome c peroxidase is a mitochondrial heme-based H2O2 sensor that modulates antioxidant defense[J]. Free Radical Biology and Medicine, 2013, 65: 541-551. |
| 6 | HIGUCHI T. Microbial degradation of lignin: role of lignin peroxidase, manganese peroxidase, and laccase[J]. Proceedings of the Japan Academy, Series B, 2004, 80(5): 204-214. |
| 7 | SIGOILLOT J C, BERRIN J G, BEY M, et al. Fungal strategies for lignin degradation[M/OL]//Lignins: biosynthesis, biodegradation and bioengineering. Amsterdam: Elsevier, 2012: 263-308. (2012-06-08)[2024-03-01]. . |
| 8 | COSIO C, DUNAND C. Specific functions of individual class Ⅲ peroxidase genes[J]. Journal of Experimental Botany, 2009, 60(2): 391-408. |
| 9 | PASSARDI F, PENEL C, DUNAND C. Performing the paradoxical: how plant peroxidases modify the cell wall[J]. Trends in Plant Science, 2004, 9(11): 534-540. |
| 10 | BARCELÓ A R, POMAR F. Oxidation of cinnamyl alcohols and aldehydes by a basic peroxidase from lignifying Zinnia elegans hypocotyls[J]. Phytochemistry, 2001, 57(7): 1105-1113. |
| 11 | BERNARDS M A, FLEMING W D, LLEWELLYN D B, et al. Biochemical characterization of the suberization-associated anionic peroxidase of potato[J]. Plant Physiology, 1999, 121(1): 135-146. |
| 12 | GAZARYAN I G, LAGRIMINI L M, ASHBY G A, et al. Mechanism of indole-3-acetic acid oxidation by plant peroxidases: anaerobic stopped-flow spectrophotometric studies on horseradish and tobacco peroxidases[J]. The Biochemical Journal, 1996, 313 (3): 841-847. |
| 13 | ALLISON S D, SCHULTZ J C. Differential activity of peroxidase isozymes in response to wounding, gypsy moth, and plant hormones in northern red oak (Quercus rubra L.) [J]. Journal of Chemical Ecology, 2004, 30(7): 1363-1379. |
| 14 | LISZKAY A, KENK B, SCHOPFER P. Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth[J]. Planta, 2003, 217(4): 658-667. |
| 15 | MCINNIS S M, DESIKAN R, HANCOCK J T, et al. Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: potential signalling crosstalk? [J]. The New Phytologist, 2006, 172(2): 221-228. |
| 16 | HUANG R H, XIA R X, HU L M, et al. Antioxidant activity and oxygen-scavenging system in orange pulp during fruit ripening and maturation[J]. Scientia Horticulturae, 2007, 113(2): 166-172. |
| 17 | PANDEY V P, AWASTHI M, SINGH S, et al. A comprehensive review on function and application of plant peroxidases[J]. Biochemistry & Analytical Biochemistry, 2017, 6(1): 308. |
| 18 | LIN Z L, THORSEN T, ARNOLD F H. Functional expression of horseradish peroxidase in E. coli by directed evolution[J]. Biotechnology Progress, 1999, 15(3): 467-471. |
| 19 | ASAD S, DABIRMANESH B, GHAEMI N, et al. Studies on the refolding process of recombinant horseradish peroxidase[J]. Molecular Biotechnology, 2013, 54(2): 484-492. |
| 20 | GUNDINGER T, SPADIUT O. A comparative approach to recombinantly produce the plant enzyme horseradish peroxidase in Escherichia coli [J]. Journal of Biotechnology, 2017, 248: 15-24. |
| 21 | CHAUHAN S, KANG T J. Soluble expression of horseradish peroxidase in Escherichia coli and its facile activation[J]. Journal of Bioscience and Bioengineering, 2018, 126(4): 431-435. |
| 22 | WANG N, REN K, JIA R, et al. Expression of a fungal manganese peroxidase in Escherichia coli: a comparison between the soluble and refolded enzymes[J]. BMC Biotechnology, 2016, 16(1): 87. |
| 23 | LIN M I, NAGATA T, KATAHIRA M. High yield production of fungal manganese peroxidases by E. coli through soluble expression, and examination of the activities[J]. Protein Expression and Purification, 2018, 145: 45-52. |
| 24 | ALFI A, ZHU B, DAMNJANOVIĆ J, et al. Production of active manganese peroxidase in Escherichia coli by co-expression of chaperones and in vitro maturation by ATP-dependent chaperone release[J]. Journal of Bioscience and Bioengineering, 2019, 128(3): 290-295. |
| 25 | GU L N, LAJOIE C, KELLY C. Expression of a Phanerochaete chrysosporium manganese peroxidase gene in the yeast Pichia pastoris [J]. Biotechnology Progress, 2003, 19(5): 1403-1409. |
| 26 | MORAWSKI B, LIN Z L, CIRINO P, et al. Functional expression of horseradish peroxidase in Saccharomyces cerevisiae and Pichia pastoris [J]. Protein Engineering, 2000, 13(5): 377-384. |
| 27 | MORAWSKI B, QUAN S, ARNOLD F H. Functional expression and stabilization of horseradish peroxidase by directed evolution in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2001, 76(2): 99-107. |
| 28 | GMEINER C, SPADIUT O. Effects of different media supplements on the production of an active recombinant plant peroxidase in a Pichia pastoris Δoch1 strain[J]. Bioengineered, 2015, 6(3): 175-178. |
| 29 | KRAINER F W, DARNHOFER B, BIRNER-GRUENBERGER R, et al. Recombinant production of a peroxidase-protein G fusion protein in Pichia pastoris [J]. Journal of Biotechnology, 2016, 219: 24-27. |
| 30 | KRAINER F W, GERSTMANN M A, DARNHOFER B, et al. Biotechnological advances towards an enhanced peroxidase production in Pichia pastoris [J]. Journal of Biotechnology, 2016, 233: 181-189. |
| 31 | CONESA A, JEENES D, ARCHER D B, et al. Calnexin overexpression increases manganese peroxidase production in Aspergillus niger [J]. Applied and Environmental Microbiology, 2002, 68(2): 846-851. |
| 32 | STEWART P, WHITWAM R E, KERSTEN P J, et al. Efficient expression of a Phanerochaete chrysosporium manganese peroxidase gene in Aspergillus oryzae [J]. Applied and Environmental Microbiology, 1996, 62(3): 860-864. |
| 33 | KAUR J, KUMAR A, KAUR J. Strategies for optimization of heterologous protein expression in E. coli: roadblocks and reinforcements[J]. International Journal of Biological Macromolecules, 2018, 106: 803-822. |
| 34 | WANG Y T, HAN H, CUI B Q, et al. A glutathione peroxidase from Antarctic psychrotrophic bacterium Pseudoalteromonas sp. ANT506: cloning and heterologous expression of the gene and characterization of recombinant enzyme[J]. Bioengineered, 2017, 8(6): 742-749. |
| 35 | LIAO J, WANG K Y, YAO W R, et al. Cloning, expression and antioxidant activity of a thioredoxin peroxidase from Branchiostoma belcheri tsingtaunese [J]. PLoS One, 2017, 12(4): e0175162. |
| 36 | KHAN S I, ZADA N S, SAHINKAYA M, et al. Cloning, expression and biochemical characterization of lignin-degrading DyP-type peroxidase from Bacillus sp. Strain BL5[J]. Enzyme and Microbial Technology, 2021, 151: 109917. |
| 37 | CHEN W T, ZHENG L L, JIA R, et al. Cloning and expression of a new manganese peroxidase from Irpex lacteus F17 and its application in decolorization of reactive black 5[J]. Process Biochemistry, 2015, 50(11): 1748-1759. |
| 38 | FATTAHIAN Y, RIAHI-MADVAR A, MIRZAEE R, et al. Heterologous expression, purification and characterization of a peroxidase isolated from Lepidium draba [J]. The Protein Journal, 2017, 36(6): 461-471. |
| 39 | GRIGORENKO V G, ANDREEVA I P, RUBTSOVA M Y, et al. Recombinant horseradish peroxidase: production and analytical applications[J]. Biochemistry (Moscow), 2015, 80(4): 408-416. |
| 40 | YIN J C, LI G X, REN X F, et al. Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes[J]. Journal of Biotechnology, 2007, 127(3): 335-347. |
| 41 | PECH-CANUL A C, CARRILLO-CAMPOS J, BALLINAS-CASARRUBIAS M L, et al. Functional expression and one-step protein purification of manganese peroxidase 1 (rMnP1) from Phanerochaetechrysosporium using the E. coli-expression system[J]. International Journal of Molecular Sciences, 2020, 21(2): 416. |
| 42 | XU H, GUO M Y, GAO Y H, et al. Expression and characteristics of manganese peroxidase from Ganoderma lucidum in Pichia pastoris and its application in the degradation of four dyes and phenol[J]. BMC Biotechnology, 2017, 17(1): 19. |
| 43 | WANG W, WEN X H. Expression of lignin peroxidase H2 from Phanerochaete chrysosporium by multi-copy recombinant Pichia strain[J]. Journal of Environmental Sciences, 2009, 21(2): 218-222. |
| 44 | KIM S J, LEE J A, WON K, et al. Functional expression of Coprinus cinereus peroxidase in Pichia pastoris [J]. Process Biochemistry, 2009, 44(7): 731-735. |
| 45 | VLAMIS-GARDIKAS A, SMITH A T, CLEMENTS J M, et al. Expression of active horseradish peroxidase in Saccharomyces cerevisiae [J]. Biochemical Society Transactions, 1992, 20(2): 111S. |
| 46 | KRAINER F W, GMEINER C, NEUTSCH L, et al. Knockout of an endogenous mannosyltransferase increases the homogeneity of glycoproteins produced in Pichia pastoris [J]. Scientific Reports, 2013, 3: 3279. |
| 47 | SU X Y, SCHMITZ G, ZHANG M L, et al. Heterologous gene expression in filamentous fungi[J]. Advances in Applied Microbiology, 2012, 81: 1-61. |
| 48 | CONESA A, VAN DEN HONDEL C A, PUNT P J. Studies on the production of fungal peroxidases in Aspergillus niger [J]. Applied and Environmental Microbiology, 2000, 66(7): 3016-3023. |
| 49 | CONESA A, VAN DE VELDE F, VAN RANTWIJK F, et al. Expression of the Caldariomyces fumago chloroperoxidase in Aspergillus niger and characterization of the recombinant enzyme[J]. The Journal of Biological Chemistry, 2001, 276(21): 17635-17640. |
| 50 | SHAKERI M, SUGANO Y, SHODA M. Stable repeated-batch production of recombinant dye-decolorizing peroxidase (rDyP) from Aspergillus oryzae [J]. Journal of Bioscience and Bioengineering, 2008, 105(6): 683-686. |
| 51 | MAYFIELD M B, KISHI K, ALIC M, et al. Homologous expression of recombinant manganese peroxidase in Phanerochaete chrysosporium [J]. Applied and Environmental Microbiology, 1994, 60(12): 4303-4309. |
| 52 | IRIE T, HONDA Y, WATANABE T, et al. Homologous expression of recombinant manganese peroxidase genes in ligninolytic fungus Pleurotus ostreatus [J]. Applied Microbiology and Biotechnology, 2001, 55(5): 566-570. |
| 53 | KAMEI I, TOMITAKA N, MOTODA T, et al. Selective homologous expression of recombinant manganese peroxidase isozyme of salt-tolerant white-rot fungus Phlebia sp. MG-60, and its salt-tolerance and thermostability[J]. Journal of Microbiology and Biotechnology, 2022, 32(2): 248-255. |
| 54 | SALEHI S, ABDOLLAHI K, PANAHI R, et al. Applications of biocatalysts for sustainable oxidation of phenolic pollutants: a review[J]. Sustainability, 2021, 13(15): 8620. |
| 55 | CHATHA S A S, ASGHER M, IQBAL H M N. Enzyme-based solutions for textile processing and dye contaminant biodegradation-a review[J]. Environmental Science and Pollution Research, 2017, 24(16): 14005-14018. |
| 56 | COUTO C F, LANGE L C, AMARAL M C S. Occurrence, fate and removal of pharmaceutically active compounds (PhACs) in water and wastewater treatment plants — a review[J]. Journal of Water Process Engineering, 2019, 32: 100927. |
| 57 | SELLAMI K, COUVERT A, NASRALLAH N, et al. Peroxidase enzymes as green catalysts for bioremediation and biotechnological applications: a review[J]. The Science of the Total Environment, 2022, 806(Pt 2): 150500. |
| 58 | BILAL M, RASHEED T, IQBAL H M N, et al. Peroxidases-assisted removal of environmentally-related hazardous pollutants with reference to the reaction mechanisms of industrial dyes[J]. Science of the Total Environment, 2018, 644: 1-13. |
| 59 | AZIZI A, ABOUSEOUD M, AHMEDI A. Phenol removal by soluble and alginate entrapped turnip peroxidase[J]. Journal of Biochemical Technology, 2014, 5: 795-800. |
| 60 | AZIZI A, ABOUSEOUD M, AMRANE A. Phenol removal by a sequential combined Fenton-enzymatic process[J]. Nature Environment & Pollution Technology, 2017, 16(1): 321-330. |
| 61 | ALEMZADEH I, NEJATI S. Phenols removal by immobilized horseradish peroxidase[J]. Journal of Hazardous Materials, 2009, 166(2/3): 1082-1086. |
| 62 | VASILEVA N, GODJEVARGOVA T, IVANOVA D, et al. Application of immobilized horseradish peroxidase onto modified acrylonitrile copolymer membrane in removing of phenol from water[J]. International Journal of Biological Macromolecules, 2009, 44(2): 190-194. |
| 63 | SELLAMI K, COUVERT A, NASRALLAH N, et al. Bio-based and cost-effective method for phenolic compounds removal using cross-linked enzyme aggregates[J]. Journal of Hazardous Materials, 2021, 403: 124021. |
| 64 | BAYRAMOĞLU G, ARıCA M Y. Enzymatic removal of phenol and p-chlorophenol in enzyme reactor: horseradish peroxidase immobilized on magnetic beads[J]. Journal of Hazardous Materials, 2008, 156(1-3): 148-155. |
| 65 | BESHARATI VINEH M, SABOURY A A, POOSTCHI A A, et al. Physical adsorption of horseradish peroxidase on reduced graphene oxide nanosheets functionalized by amine: a good system for biodegradation of high phenol concentration in wastewater[J]. International Journal of Environmental Research, 2018, 12(1): 45-57. |
| 66 | ASHRAF H, HUSAIN Q. Application of immobilized peroxidase for the removal of p-bromophenol from polluted water in batch and continuous processes[J]. Journal of Water Reuse and Desalination, 2011, 1(1): 52-60. |
| 67 | PETRONIJEVIĆ M, PANIĆ S, SAVIĆ S, et al. Characterization and application of biochar-immobilized crude horseradish peroxidase for removal of phenol from water[J]. Colloids and Surfaces B, Biointerfaces, 2021, 208: 112038. |
| 68 | EL-NAGGAR M E, ABDEL-ATY A M, WASSEL A R, et al. Immobilization of horseradish peroxidase on cationic microporous starch: physico-bio-chemical characterization and removal of phenolic compounds[J]. International Journal of Biological Macromolecules, 2021, 181: 734-742. |
| 69 | PANTIĆ N, PRODANOVIĆ R, ĐURĐIĆ K I, et al. Optimization of phenol removal with horseradish peroxidase encapsulated within tyramine-alginate micro-beads[J]. Environmental Technology & Innovation, 2021, 21: 101211. |
| 70 | BILAL M, RASHEED T, IQBAL H M N, et al. Horseradish peroxidase immobilization by copolymerization into cross-linked polyacrylamide gel and its dye degradation and detoxification potential[J]. International Journal of Biological Macromolecules, 2018, 113: 983-990. |
| 71 | WEBER A C, SILVA B E DA, CORDEIRO S G, et al. Immobilization of commercial horseradish peroxidase in calcium alginate-starch hybrid support and its application in the biodegradation of phenol red dye[J]. International Journal of Biological Macromolecules, 2023, 246: 125723. |
| 72 | PARVEEN S, ASGHER M, BILAL M. Lignin peroxidase-based cross-linked enzyme aggregates (LiP-CLEAs) as robust biocatalytic materials for mitigation of textile dyes-contaminated aqueous solution[J]. Environmental Technology & Innovation, 2021, 21: 101226. |
| 73 | ALI L, ALGAITHI R, HABIB H M, et al. Soybean peroxidase-mediated degradation of an azo dye-a detailed mechanistic study[J]. BMC Biochemistry, 2013, 14: 35. |
| 74 | ZHANG Y R, REN J, WANG Q, et al. Oxidation characteristics and degradation potential of a dye-decolorizing peroxidase from Bacillus amyloliquefaciens for crystal violet dye[J]. Biochemical Engineering Journal, 2021, 168: 107930. |
| 75 | BOUCHERIT N, ABOUSEOUD M, ADOUR L. Degradation of direct azo dye by Cucurbita pepo free and immobilized peroxidase[J]. Journal of Environmental Sciences, 2013, 25(6): 1235-1244. |
| 76 | WANG H L, LI P, PANG M, et al. Rapid decolourization of azo dyes by a new isolated higher manganese peroxidase producer: Phanerochaete sp. HSD[J]. Biochemical Engineering Journal, 2009, 46(3): 327-333. |
| 77 | KALSOOM U, ASHRAF S S, MEETANI M A, et al. Mechanistic study of a diazo dye degradation by Soybean Peroxidase[J]. Chemistry Central Journal, 2013, 7(1): 93. |
| 78 | PREETHI P S, VICKRAM S, DAS R, et al. Bioprospecting of novel peroxidase from Streptomyces coelicolor strain SPR7 for carcinogenic azo dyes decolorization[J]. Chemosphere, 2023, 310: 136836. |
| 79 | HAMDI S, ALLALA F, MECHRI S, et al. Biochemical and molecular characterization of a new heme peroxidase from Aspergillus niger CTM10002, and its application in textile reactive dye decolorization[J]. Process Biochemistry, 2022, 121: 619-634. |
| 80 | KALSOOM U, BHATTI H N, AFTAB K, et al. Biocatalytic potential of Brassica oleracea L. var. botrytis leaves peroxidase for efficient degradation of textile dyes in aqueous medium[J]. Bioprocess and Biosystems Engineering, 2023, 46(3): 453-465. |
| 81 | AI J, ZHANG W J, LIAO G Y, et al. NH2Fe3O4@SiO2 supported peroxidase catalyzed H2O2 for degradation of endocrine disrupter from aqueous solution: roles of active radicals and NOMs[J]. Chemosphere, 2017, 186: 733-742. |
| 82 | TABOADA-PUIG R, EIBES G, LLORET L, et al. Fostering the action of versatile peroxidase as a highly efficient biocatalyst for the removal of endocrine disrupting compounds[J]. New Biotechnology, 2016, 33(1): 187-195. |
| 83 | MASHHADI N, TAYLOR K E, JIMENEZ N, et al. Removal of selected pharmaceuticals and personal care products from wastewater using soybean peroxidase[J]. Environmental Management, 2019, 63(3): 408-415. |
| 84 | AL-MAQDI K A, HISAINDEE S, RAUF M A, et al. Detoxification and degradation of sulfamethoxazole by soybean peroxidase and UV+ H2O2 remediation approaches[J]. Chemical Engineering Journal, 2018, 352: 450-458. |
| 85 | PYLYPCHUK I V, DANIEL G, KESSLER V G, et al. Removal of diclofenac, paracetamol, and carbamazepine from model aqueous solutions by magnetic sol-gel encapsulated horseradish peroxidase and lignin peroxidase composites[J]. Nanomaterials, 2020, 10(2): 282. |
| 86 | WEN X H, JIA Y N, LI J X. Enzymatic degradation of tetracycline and oxytetracycline by crude manganese peroxidase prepared from Phanerochaete chrysosporium [J]. Journal of Hazardous Materials, 2010, 177(1-3): 924-928. |
| 87 | ZDARTA J, DEGÓRSKA O, JANKOWSKA K, et al. Removal of persistent sulfamethoxazole and carbamazepine from water by horseradish peroxidase encapsulated into poly (vinyl chloride) electrospun fibers[J]. International Journal of Molecular Sciences, 2021, 23(1): 272. |
| 88 | GARCÍA-ZAMORA J L, LEÓN-AGUIRRE K, QUIROZ-MORALES R, et al. Chloroperoxidase-mediated halogenation of selected pharmaceutical micropollutants[J]. Catalysts, 2018, 8(1): 32. |
| 89 | HAMID M, KHALIL-UR-REHMAN. Potential applications of peroxidases[J]. Food Chemistry, 2009, 115(4): 1177-1186. |
| 90 | LINDGREN A, EMNÉUS J, RUZGAS T, et al. Amperometric detection of phenols using peroxidase-modified graphite electrodes[J]. Analytica Chimica Acta, 1997, 347(1-2): 51-62. |
| 91 | TKÁC J, STURDÍK E, GEMEINER P. Novel glucose non-interference biosensor for lactose detection based on galactose oxidase-peroxidase with and without co-immobilised β-galactosidase[J]. The Analyst, 2000, 125(7): 1285-1289. |
| 92 | GILLE D, WALTHER B, BADERTSCHER R, et al. Detection of lactose in products with low lactose content[J]. International Dairy Journal, 2018, 83: 17-19. |
| 93 | LIANG X, CHEN Y X, WEN K, et al. Urate oxidase loaded in PCN-222(Fe) with peroxidase-like activity for colorimetric detection of uric acid[J]. Journal of Materials Chemistry B, 2021, 9(34): 6811-6817. |
| 94 | AKYILMAZ E, SEZGINTÜRK M K, DINÇKAYA E. A biosensor based on urate oxidase-peroxidase coupled enzyme system for uric acid determination in urine[J]. Talanta, 2003, 61(2): 73-79. |
| 95 | SAA L, CORONADO-PUCHAU M, PAVLOV V, et al. Enzymatic etching of gold nanorods by horseradish peroxidase and application to blood glucose detection[J]. Nanoscale, 2014, 6(13): 7405-7409. |
| 96 | CSÖREGI E, GORTON L, MARKO-VARGA G. Amperometric microbiosensors for detection of hydrogen peroxide and glucose based on peroxidase-modified carbon fibers[J]. Electroanalysis, 1994, 6(11-12): 925-933. |
| 97 | FOSSATI P, PRENCIPE L. Serum triglycerides determined colorimetrically with an enzyme that produces hydrogen peroxide[J]. Clinical Chemistry, 1982, 28(10): 2077-2080. |
| 98 | PUNDIR C S, AGGARWAL V. Amperometric triglyceride bionanosensor based on nanoparticles of lipase, glycerol kinase, glycerol-3-phosphate oxidase[J]. Analytical Biochemistry, 2017, 517: 56-63. |
| 99 | ZHAO M Y, LI Y, MA X J, et al. Adsorption of cholesterol oxidase and entrapment of horseradish peroxidase in metal-organic frameworks for the colorimetric biosensing of cholesterol[J]. Talanta, 2019, 200: 293-299. |
| 100 | SINGH S, SOLANKI P R, PANDEY M K, et al. Cholesterol biosensor based on cholesterol esterase, cholesterol oxidase and peroxidase immobilized onto conducting polyaniline films[J]. Sensors and Actuators B: Chemical, 2006, 115(1): 534-541. |
| 101 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs from 1981 to 2014[J]. Journal of Natural Products, 2016, 79(3): 629-661. |
| 102 | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 103 | AJIKUMAR P K, XIAO W H, TYO K E, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli [J]. Science, 2010, 330(6000): 70-74. |
| 104 | OGUCHI T, TAWAKI S I, UYAMA H, et al. Soluble polyphenol[J]. Macromolecular rapid communications, 1999, 20(7): 401-403. |
| 105 | UYAMA H, KURIOKA H, SUGIHARA J, et al. Enzymatic synthesis and thermal properties of a new class of polyphenol[J]. Bulletin of the Chemical Society of Japan, 1996, 69(1): 189-193. |
| 106 | TONAMI H, UYAMA H, KOBAYASHI S, et al. Peroxidase-catalyzed oxidative polymerization of m-substituted phenol derivatives[J]. Macromolecular Chemistry and Physics, 1999, 200(10): 2365-2371. |
| 107 | DUBEY S, SINGH D, MISRA R A. Enzymatic synthesis and various properties of poly(catechol)[J]. Enzyme and Microbial Technology, 1998, 23(7-8): 432-437. |
| 108 | TANG B, WANG Y, LIANG H L, et al. Studies on the oxidation reaction of tyrosine (Tyr) with H2O2 catalyzed by horseradish peroxidase (HRP) in alcohol-water medium by spectrofluorimetry and differential spectrophotometry[J]. Spectrochimica Acta Part A, Molecular and Biomolecular Spectroscopy, 2006, 63(3): 609-613. |
| 109 | SALIU F, TOLPPA E L, ZOIA L, et al. Horseradish peroxidase catalyzed oxidative cross-coupling reactions: the synthesis of ‘unnatural’ dihydrobenzofuran lignans[J]. Tetrahedron Letters, 2011, 52(30): 3856-3860. |
| 110 | LI C, LU J, XU X F, et al. pH-switched HRP-catalyzed dimerization of resveratrol: a selective biomimetic synthesis[J]. Green Chemistry, 2012, 14(12): 3281-3284. |
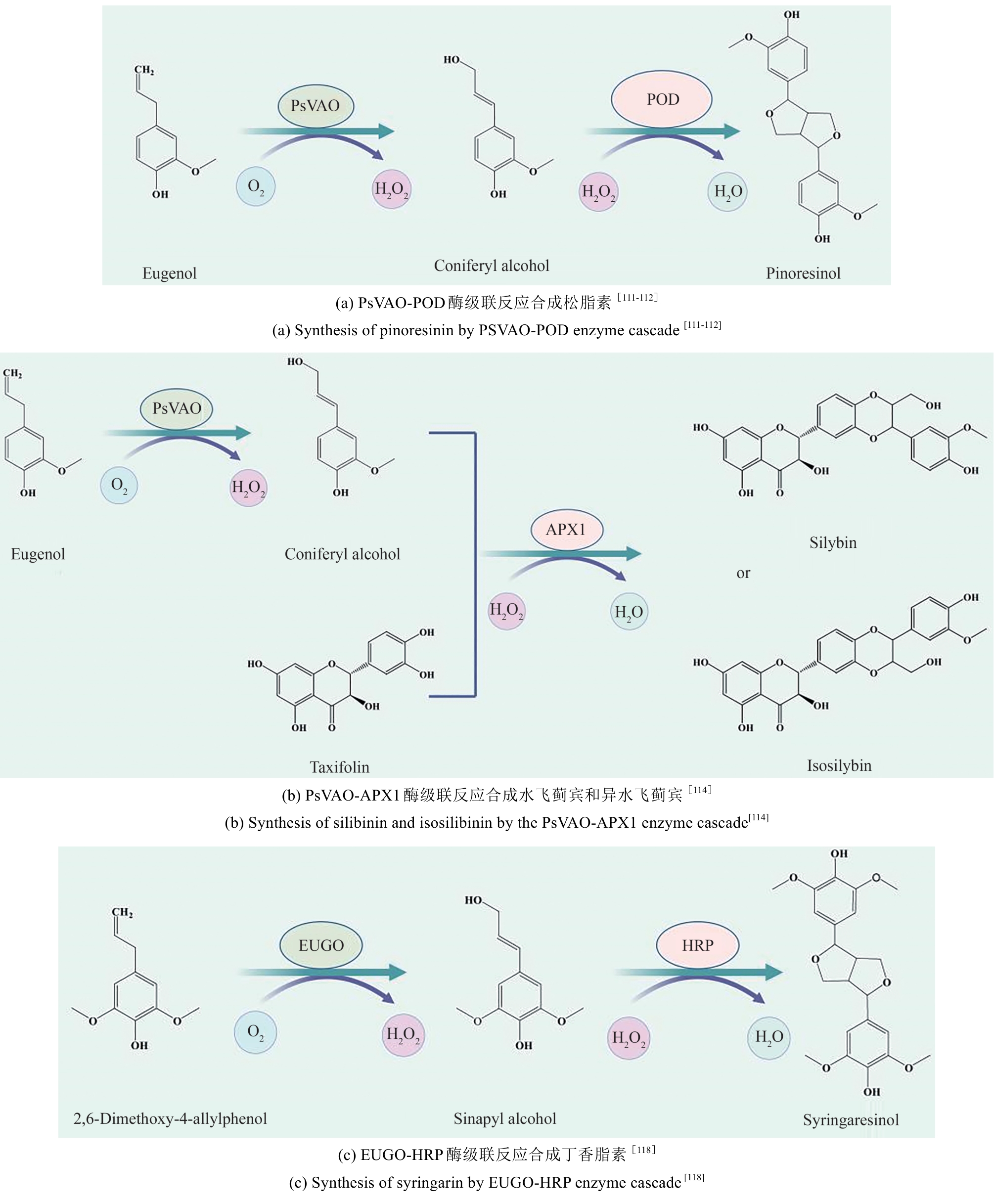
| 111 | RICKLEFS E, GIRHARD M, KOSCHORRECK K, et al. Two-step one-pot synthesis of pinoresinol from eugenol in an enzymatic cascade[J]. ChemCatChem, 2015, 7(12): 1857-1864. |
| 112 | LÜ Y K, CHENG X Z, DU G C, et al. Engineering of an H2O2 auto-scavenging in vivo cascade for pinoresinol production[J]. Biotechnology and Bioengineering, 2017, 114(9): 2066-2074. |
| 113 | ABENAVOLI L, CAPASSO R, MILIC N, et al. Milk thistle in liver diseases: past, present, future[J]. Phytotherapy Research, 2010, 24(10): 1423-1432. |
| 114 | LÜ Y K, XU S, LYU Y B, et al. Engineering enzymatic cascades for the efficient biotransformation of eugenol and taxifolin to silybin and isosilybin[J]. Green Chemistry, 2019, 21(7): 1660-1667. |
| 115 | YANG J Z, LIANG J C, SHAO L, et al. Green production of silybin and isosilybin by merging metabolic engineering approaches and enzymatic catalysis[J]. Metabolic Engineering, 2020, 59: 44-52. |
| 116 | LI X L, ZHOU Z, LI W N, et al. Design of stable and self-regulated microbial consortia for chemical synthesis[J]. Nature Communications, 2022, 13(1): 1554. |
| 117 | PARK S Y, YANG D, HA S H, et al. Production of phenylpropanoids and flavonolignans from glycerol by metabolically engineered Escherichia coli [J]. Biotechnology and Bioengineering, 2022, 119(3): 946-962. |
| 118 | HABIB M, TRAJKOVIC M, FRAAIJE M W. The biocatalytic synthesis of syringaresinol from 2,6-dimethoxy-4-allylphenol in one-pot using a tailored oxidase/peroxidase system[J]. ACS Catalysis, 2018, 8(6): 5549-5552. |
| 119 | GUO Y M, ALVIGINI L, SAIFUDDIN M, et al. One-pot biocatalytic synthesis of rac-syringaresinol from a lignin-derived phenol[J]. ACS Catalysis, 2023, 13(22): 14639-14649. |
| [1] | LIU Kuanqing, ZHANG Yi-Heng P.Job. Biological degradation and utilization of lignin [J]. Synthetic Biology Journal, 2024, 5(6): 1264-1278. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||