Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (2): 161-180.DOI: 10.12211/2096-8280.2020-087
• Invited Review • Previous Articles Next Articles
Synthetic biology boosts biological depolymerization and upgrading of waste plastics
QIAN Xiujuan1, LIU Jiawei1, XUE Rui1, LIU Haojie1, WEN Xiaohong1, YANG Lu1, XU Anming1, XU Bin1, XIN Fengxue1,2, ZHOU Jie1,2, DONG Weiliang1,2, JIANG Min1,2
- 1.College of Biotechnology and Pharmaceutical Engineering,Nanjing Tech University,Nanjing 211816,Jiangsu,China
2.State Key Laboratory of Materials-Oriented Chemical Engineering,Nanjing Tech University,Nanjing 211816,Jiangsu,China
-
Received:2020-12-04Revised:2021-02-11Online:2021-04-30Published:2021-04-30 -
Contact:DONG Weiliang, JIANG Min
合成生物学助力废弃塑料资源生物解聚与升级再造
钱秀娟1, 刘嘉唯1, 薛瑞1, 刘豪杰1, 闻小红1, 杨璐1, 徐安明1, 许斌1, 信丰学1,2, 周杰1,2, 董维亮1,2, 姜岷1,2
- 1.南京工业大学生物与制药工程学院,江苏 南京 211816
2.南京工业大学材料化学工程国家重点实验室,江苏 南京 211816
-
通讯作者:董维亮,姜岷 -
作者简介:钱秀娟 (1992—),女,博士,博士后,研究方向为代谢工程及合成生物学。E-mail:xiujuanqian@njtech.edu.cn
董维亮(1988—),男,博士,教授,研究方向为环境污染物的生物降解与转化利用。E-mail:dwl@njtech.edu.cn
姜岷(1972—),男,博士,教授,研究方向为废弃碳资源利用人工多细胞体系设计与构建。E-mail:jiangmin@njtech.edu.cn -
基金资助:国家自然科学基金国际(地区)合作与交流项目(31961133017);国家重点研发计划“合成生物学”重点专项(2019YFA0905500);国家自然科学基金(21978129);江苏省农业自主创新计划(CX(19)3104);江苏省研究生科研与实践创新计划(KYCX20_1100)
CLC Number:
Cite this article
QIAN Xiujuan, LIU Jiawei, XUE Rui, LIU Haojie, WEN Xiaohong, YANG Lu, XU Anming, XU Bin, XIN Fengxue, ZHOU Jie, DONG Weiliang, JIANG Min. Synthetic biology boosts biological depolymerization and upgrading of waste plastics[J]. Synthetic Biology Journal, 2021, 2(2): 161-180.
钱秀娟, 刘嘉唯, 薛瑞, 刘豪杰, 闻小红, 杨璐, 徐安明, 许斌, 信丰学, 周杰, 董维亮, 姜岷. 合成生物学助力废弃塑料资源生物解聚与升级再造[J]. 合成生物学, 2021, 2(2): 161-180.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-087
| 塑料 分类 | 降解菌 | 降解温度/℃ | 降解效果 | 文献 |
|---|---|---|---|---|
| PET | Fusarium solani | 30 | PET纤维表面改性 | [ |
| Humicola insolens | 30 | PET纤维表面改性 | [ | |
| Thermobifida fusca | 30 | PET纤维表面改性 | [ | |
| Saccharomonospora viridis | 30 | PET纤维表面改性 | [ | |
| Ideonella sakaiensis 201-F6 | 30 | 6周内能完全降解低结晶度PET薄膜 | [ | |
| PU | Aspergillus flavus (ITCC 6051) | 28±2 | 30 d降解60.6%聚酯型PU薄膜 | [ |
| Aspergillus tubingensis | 37 | 14 d降解聚酯型PU薄膜成碎片 | [ | |
| Aspergillus sp. strain S45 | 37 | 28 d降解20%聚酯型PU薄膜 | [ | |
| Cladosporium pseudocladosporioides, Cladosporium tenuissimum, Cladosporium asperulatum, Cladosporium montecillanum, Aspergillus fumigatus, Penicillium chrysogenum | 25~30 | 21 d质降解10%~65%聚酯型PU薄膜 | [ |
Tab. 1 Microorganisms responsible for depolymerizing plastics through hydrolysis
| 塑料 分类 | 降解菌 | 降解温度/℃ | 降解效果 | 文献 |
|---|---|---|---|---|
| PET | Fusarium solani | 30 | PET纤维表面改性 | [ |
| Humicola insolens | 30 | PET纤维表面改性 | [ | |
| Thermobifida fusca | 30 | PET纤维表面改性 | [ | |
| Saccharomonospora viridis | 30 | PET纤维表面改性 | [ | |
| Ideonella sakaiensis 201-F6 | 30 | 6周内能完全降解低结晶度PET薄膜 | [ | |
| PU | Aspergillus flavus (ITCC 6051) | 28±2 | 30 d降解60.6%聚酯型PU薄膜 | [ |
| Aspergillus tubingensis | 37 | 14 d降解聚酯型PU薄膜成碎片 | [ | |
| Aspergillus sp. strain S45 | 37 | 28 d降解20%聚酯型PU薄膜 | [ | |
| Cladosporium pseudocladosporioides, Cladosporium tenuissimum, Cladosporium asperulatum, Cladosporium montecillanum, Aspergillus fumigatus, Penicillium chrysogenum | 25~30 | 21 d质降解10%~65%聚酯型PU薄膜 | [ |
| 塑料分类 | 降解底物 | 解聚酶来源 | 解聚酶 | 降解温度/℃ | 降解能力 | 文献 |
|---|---|---|---|---|---|---|
| PET | 饮料瓶 | Thermobifida fusca DSM43793 | TfH | 55 | 21 d质量损失50% | [ |
| 低结晶度(7%)PET薄膜 | Humicola insolens | HiC | 70 | 96 h重量损失97% | [ | |
| PET薄膜 | plant compost | LCC | 70 | 24 h重量损失25% | [ | |
| 低结晶度(1.9%)PET薄膜 | Ideonella sakaiensis 201-F6 | PETase | 30 | — | [ | |
| 低结晶度PET薄膜 | T. fusca KW3 | TfCut2 | 65~80 | 48 h重量损失12% | [ | |
| PU | Impranil DLN | Comamonas acidovorans TB-35 | PudA | 45 | — | [ |
| Impranil DLN | Pseudomonas fluorescens | PulA | 48 | — | [ | |
| Impranil DLN | Pseudomonas chlororaphis | PueA/PueB | 65/60 | — | [ | |
| 固体聚酯型PU | T. fusca KW3 | TfCut2 | 70 | 100 h重量损失1.9% | [ | |
| 固体聚酯型PU | plant compost | LCC | 70 | 100 h重量损失3.2% | [ |
Tab. 2 Depolymerases responsible for plastics depolymerization through hydrolysis
| 塑料分类 | 降解底物 | 解聚酶来源 | 解聚酶 | 降解温度/℃ | 降解能力 | 文献 |
|---|---|---|---|---|---|---|
| PET | 饮料瓶 | Thermobifida fusca DSM43793 | TfH | 55 | 21 d质量损失50% | [ |
| 低结晶度(7%)PET薄膜 | Humicola insolens | HiC | 70 | 96 h重量损失97% | [ | |
| PET薄膜 | plant compost | LCC | 70 | 24 h重量损失25% | [ | |
| 低结晶度(1.9%)PET薄膜 | Ideonella sakaiensis 201-F6 | PETase | 30 | — | [ | |
| 低结晶度PET薄膜 | T. fusca KW3 | TfCut2 | 65~80 | 48 h重量损失12% | [ | |
| PU | Impranil DLN | Comamonas acidovorans TB-35 | PudA | 45 | — | [ |
| Impranil DLN | Pseudomonas fluorescens | PulA | 48 | — | [ | |
| Impranil DLN | Pseudomonas chlororaphis | PueA/PueB | 65/60 | — | [ | |
| 固体聚酯型PU | T. fusca KW3 | TfCut2 | 70 | 100 h重量损失1.9% | [ | |
| 固体聚酯型PU | plant compost | LCC | 70 | 100 h重量损失3.2% | [ |
| 塑料分类 | 降解底物 | 降解菌 | 降解温度/℃ | 降解效果 | 文献 |
|---|---|---|---|---|---|
| PE | 改性PE膜 | Aspergillus niger M6 | 28 | 30 d质量损失20% | [ |
| 高密度PE | Arthrobacter sp. GMB5 | 30 | 30 d质量损失12% | [ | |
| 高密度PE | Pseudomonas sp. GMB7 | 30 | 30 d质量损失15% | [ | |
| 低密度PE薄膜 | Enterobacter asburiae YT1 | 30 | 60 d质量损失6.1%±0.3% | [ | |
| 低密度PE薄膜 | Bacillus sp. YP1 | 30 | 60 d质量损失10.7%±0.2% | [ | |
| PS | PS泡沫 | Mealworms (the larvae of Tenebrio molitor Linnaeus) | 30 | 30 d质量损失31.0%±1.7% | [ |
| PS薄膜 | Exiguobacterium sp. YT2 | 30 | 60 d质量损失7.4%±0.4% | [ | |
| PS薄膜 | Penicillium variabile | 24 | 16周内能矿化完 | [ |
Tab. 3 Microorganisms responsible for plastics depolymerization through non-hydrolysis pathways
| 塑料分类 | 降解底物 | 降解菌 | 降解温度/℃ | 降解效果 | 文献 |
|---|---|---|---|---|---|
| PE | 改性PE膜 | Aspergillus niger M6 | 28 | 30 d质量损失20% | [ |
| 高密度PE | Arthrobacter sp. GMB5 | 30 | 30 d质量损失12% | [ | |
| 高密度PE | Pseudomonas sp. GMB7 | 30 | 30 d质量损失15% | [ | |
| 低密度PE薄膜 | Enterobacter asburiae YT1 | 30 | 60 d质量损失6.1%±0.3% | [ | |
| 低密度PE薄膜 | Bacillus sp. YP1 | 30 | 60 d质量损失10.7%±0.2% | [ | |
| PS | PS泡沫 | Mealworms (the larvae of Tenebrio molitor Linnaeus) | 30 | 30 d质量损失31.0%±1.7% | [ |
| PS薄膜 | Exiguobacterium sp. YT2 | 30 | 60 d质量损失7.4%±0.4% | [ | |
| PS薄膜 | Penicillium variabile | 24 | 16周内能矿化完 | [ |
| 塑料分类 | 降解底物 | 解聚酶来源 | 解聚酶 | 降解温度/℃ | 降解能力 | 文献 |
|---|---|---|---|---|---|---|
| PE | 氧化后的 低密度PE | Phanerochaete chrysosporium MTCC-787 | LiP/MnP | 37 | 15 d内降解70% | [ |
| 低分子量 PE粉末 | Pseudomonas sp. E4 | alkB | 37 | 80 d内降解20% | [ | |
| 低分子量 PE粉末 | Pseudomonas aeruginosa E7 | AH系统 | 37 | 80 d内降解30% | [ | |
| PS | PS | Azotobacter beijerinckii HM121 | 非血红素氢醌 过氧化物酶 | 30 | 5 min内水解PS转 化为水溶性产物 | [ |
Tab. 4 Depolymerases responsible for plastics depolymerization through non-hydrolysis pathways
| 塑料分类 | 降解底物 | 解聚酶来源 | 解聚酶 | 降解温度/℃ | 降解能力 | 文献 |
|---|---|---|---|---|---|---|
| PE | 氧化后的 低密度PE | Phanerochaete chrysosporium MTCC-787 | LiP/MnP | 37 | 15 d内降解70% | [ |
| 低分子量 PE粉末 | Pseudomonas sp. E4 | alkB | 37 | 80 d内降解20% | [ | |
| 低分子量 PE粉末 | Pseudomonas aeruginosa E7 | AH系统 | 37 | 80 d内降解30% | [ | |
| PS | PS | Azotobacter beijerinckii HM121 | 非血红素氢醌 过氧化物酶 | 30 | 5 min内水解PS转 化为水溶性产物 | [ |
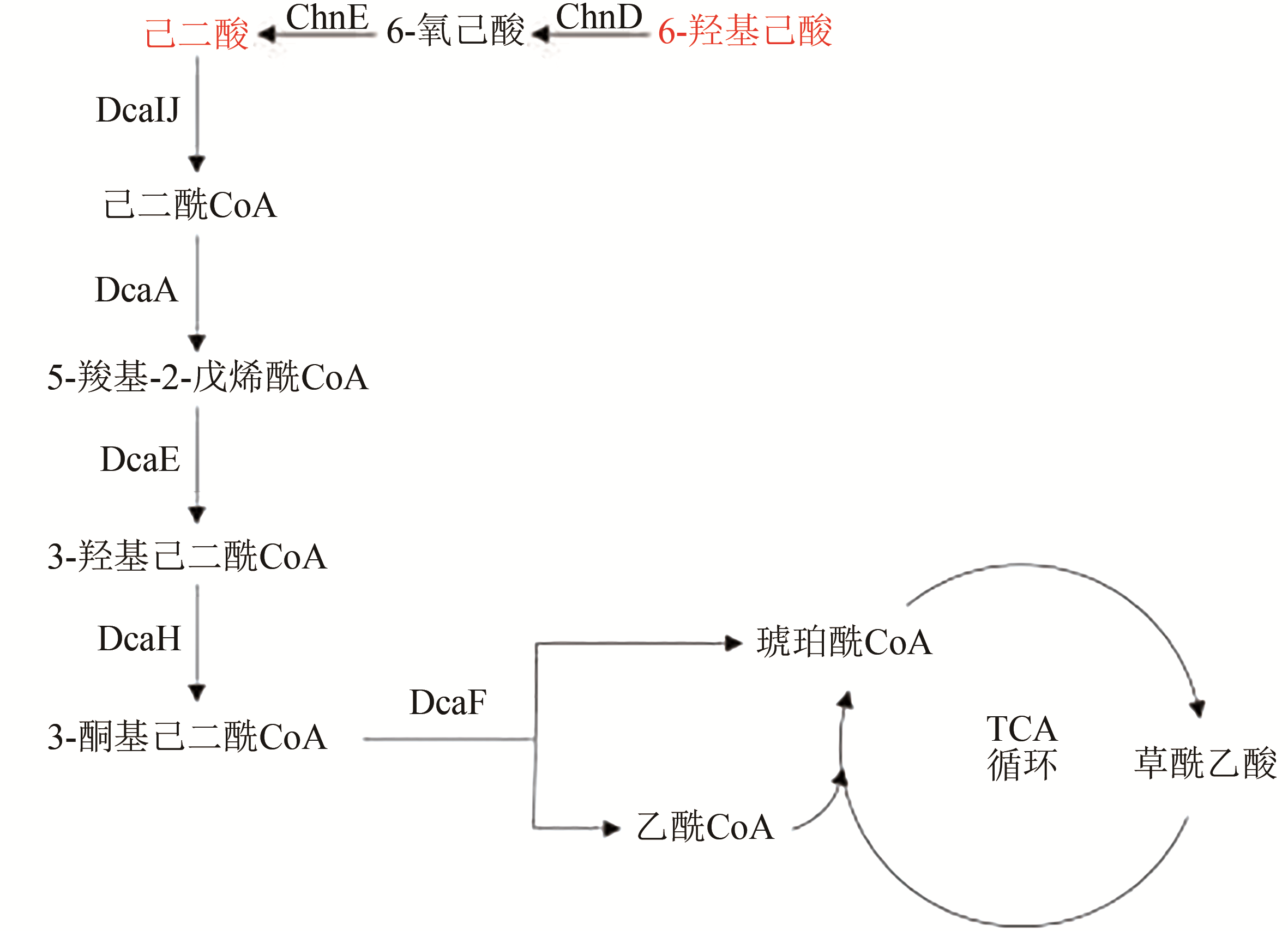
Fig. 2 Biological degradation pathway for plastics from organic acid based monomers (adipic acid, 6-hydroxyhexanoic acid, etc.)(Key enzymes in metabolic pathway: DcaIJ—succinyl-CoA transferase; DcaA—acyl-CoA dehydrogenase; DcaE—enoyl-CoA hydratase; DcaH—3-hyroxyacyl-CoA dehydrogenase; DcaF—acyl-CoA thiolase; ChnD—6-hydroxyhexanoate dehydrogenase; ChnE—6-oxohexanoate dehydrogenase)
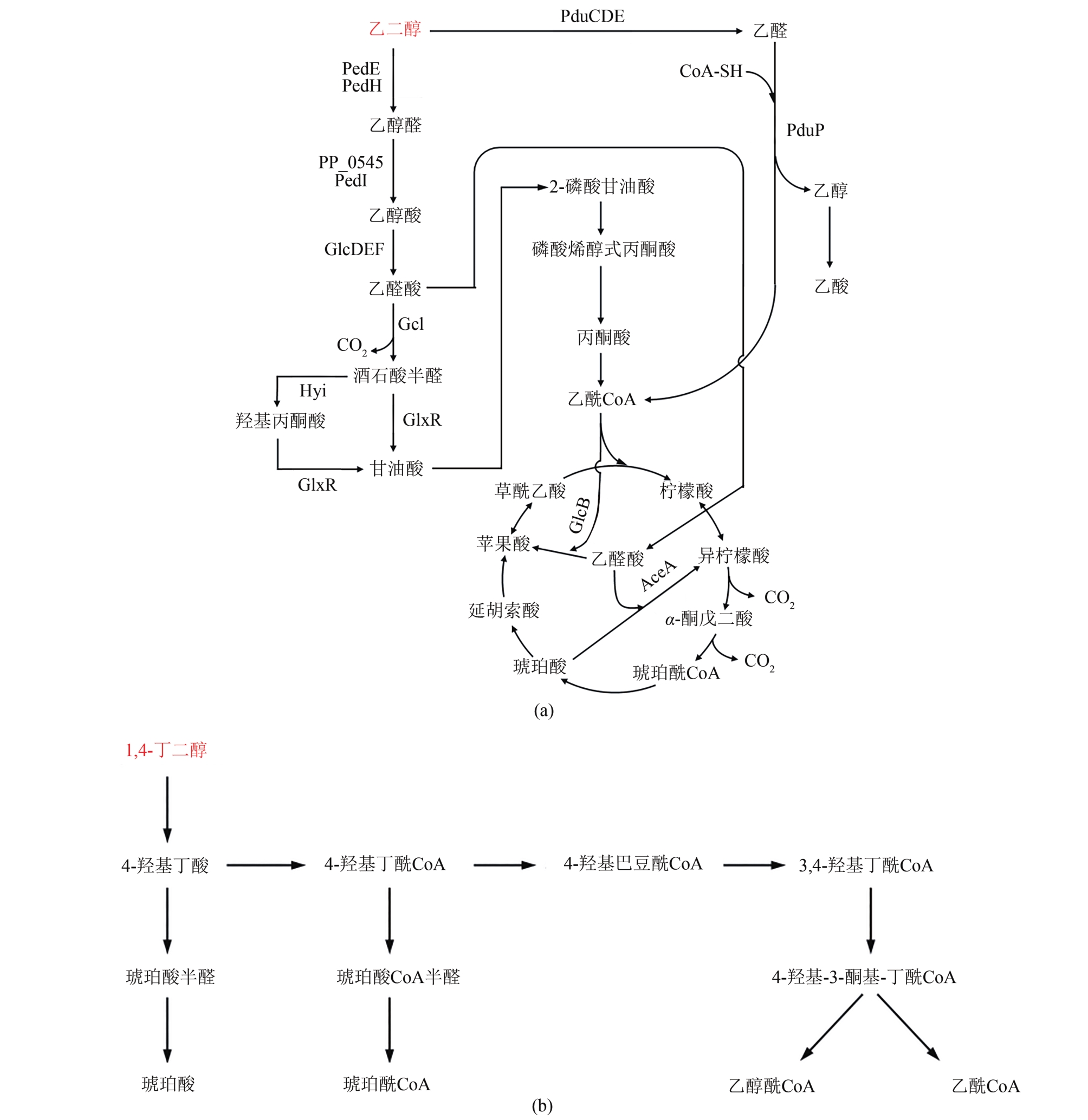
Fig. 3 Biological degradation pathway for plastic from organic alcohol based monomers (ethylene glycol, 1,4-butanediol, etc.)(Key enzymes in metabolic pathway:PedE, PedH—quinoprotein alcohol dehydrogenase; PP_0545 and PedI—aldehyde dehydrogenase; GlcDEF—glycolate oxidase; Gcl—glyoxylate carboligase; Hyi—hydroxypyruvate isomerase; GlxR—artronate semialdehyde reductase; AceA—isocitrate lyase; GlcB—malate synthase; PduP—propionaldehyde dehydrogenase)
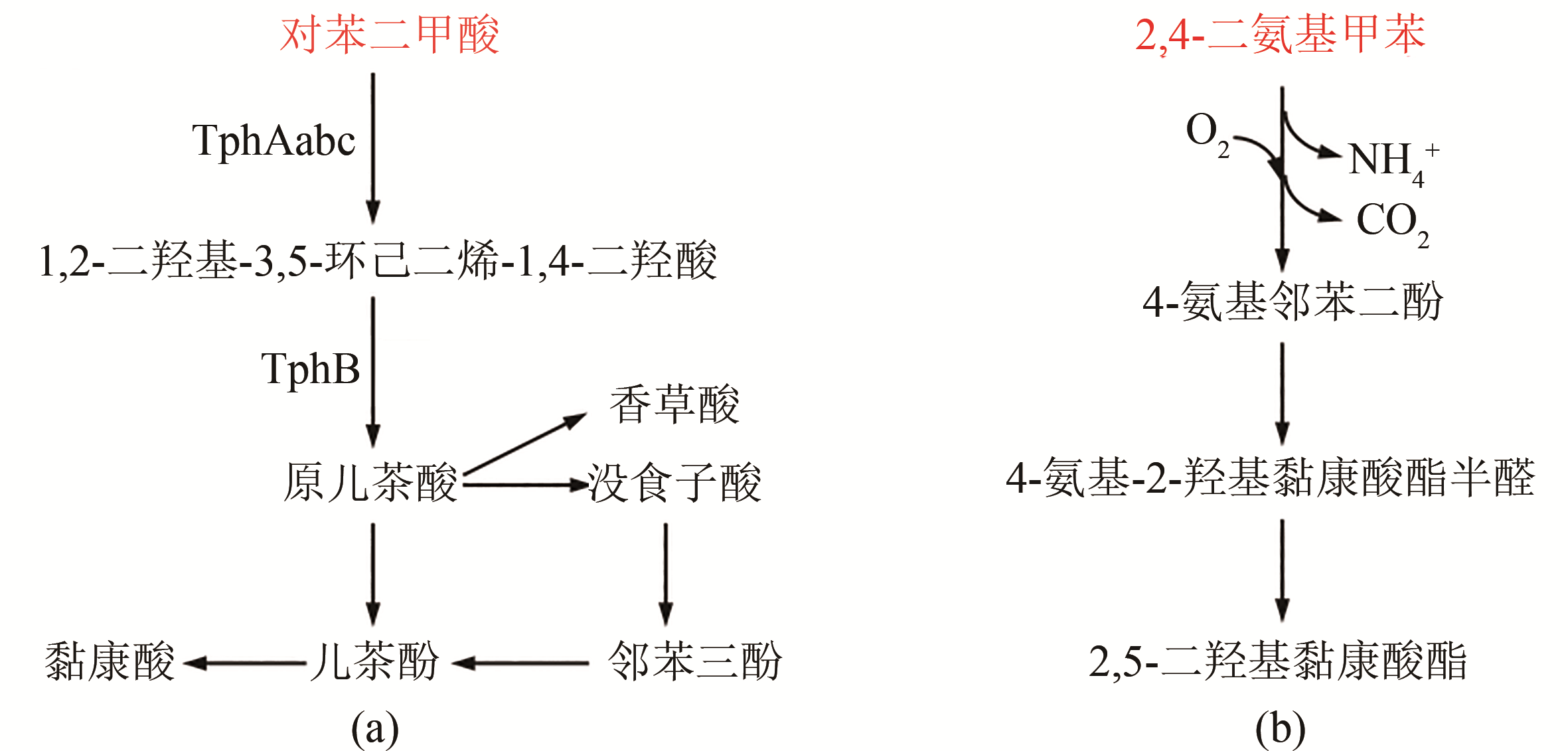
Fig. 4 Biological degradation pathway for plastics from aromatic monomers (terephthalic acid, 2,4-diaminotoluene, etc.)(Key enzymes in metabolic pathway: TphAabc—TPA 1,2-dioxygenase; TphB—1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase)
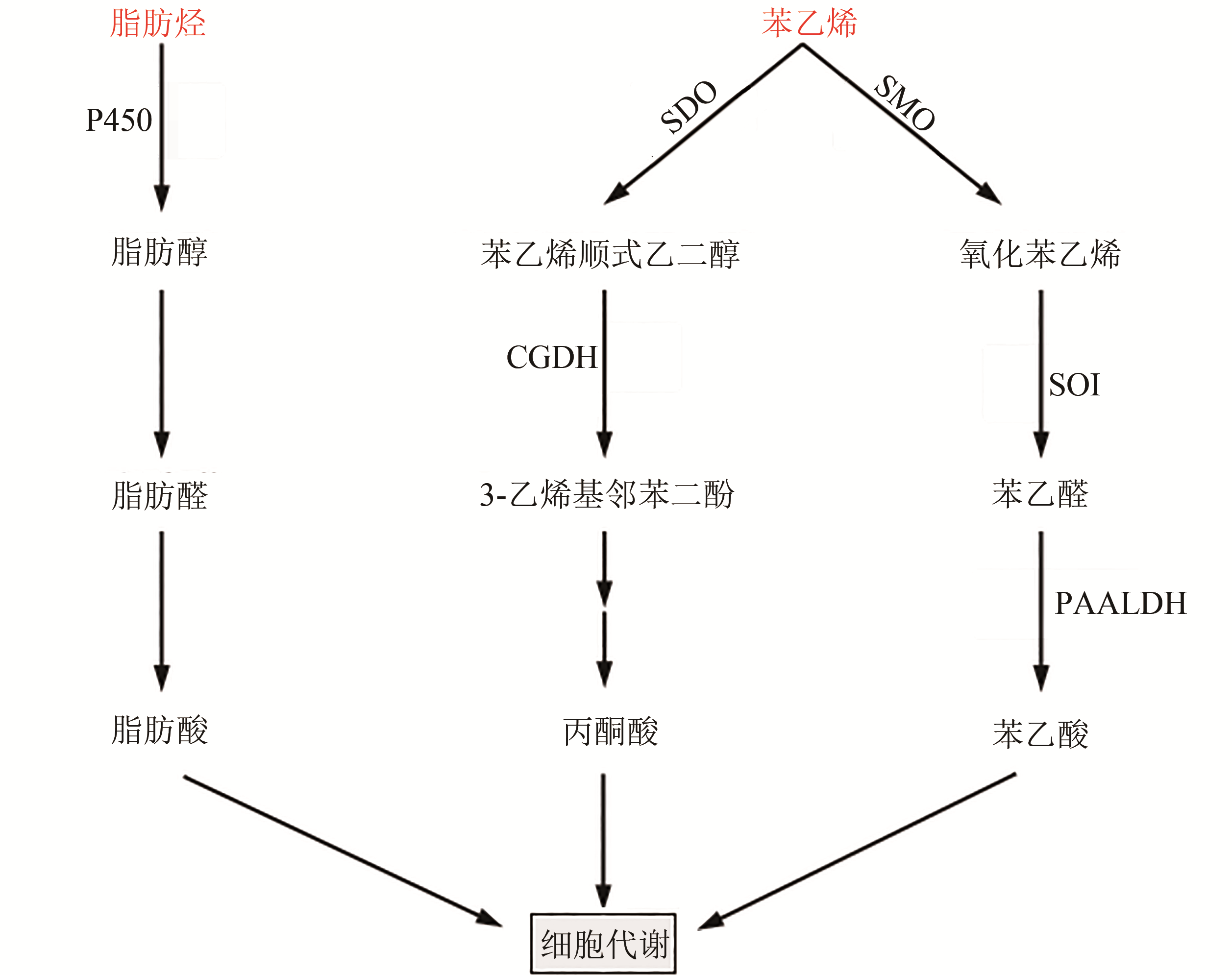
Fig. 5 Biological degradation pathway for plastics from aliphatic hydrocarbon monomers(Key enzymes in metabolic pathway: P450—monooxygenase P450; SDO—styrene dioxygenase; SMO—styrene monooxygenase; CGDH—cis-ethylene glycol dehydrogenase; SOI—styrene oxide isomerase; PAALDH—phenylacetaldehyde dehydrogenase)
| 53 | WEI Ren, ZIMMERMANN W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we?[J]. Microbial Biotechnology, 2017, 10(6): 1308-1322. |
| 54 | SANTO M, WEITSMAN R, SIVAN A. The role of the copper-binding enzyme-laccase-in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber [J]. International Biodeterioration & Biodegradation, 2013, 84: 204-210. |
| 55 | FUJISAWA m, HIRAI H, NISHIDA T. Degradation of polyethylene and Nylon-66 by the laccase-mediator system[J]. Journal of Polymers & the Environment, 2001, 9(3): 103-108. |
| 56 | ROJO F. Enzymes for aerobic degradation of alkanes[M]// TIMMIS K N. Handbook of hydrocarbon and lipid microbiology. Berlin: Springer, 2010. |
| 57 | YOON Moongyung, JEON Hyunjeong, KIM Malnam. Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell[J]. Journal of Bioremediation & Biodegradation, 2012, 3(4): 8. |
| 58 | JEON Hyunjeong, KIM Malnam. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation[J]. International Biodeterioration & Biodegradation, 2015, 103: 141-146. |
| 59 | NAKAMIYA K, SAKASITA G, OOI T, et al. Enzymatic degradation of polystyrene by hydroquinone peroxidase of Azotobacter beijerinckii HM121[J]. Journal of Fermentation & Bioengineering, 1997, 84(5): 480-482. |
| 60 | IIYOSHI Y, TSUTSUMI Y, NISHIDA T. Polyethylene degradation by lignin-degrading fungi and manganese peroxidase[J]. Journal of Wood Science, 1998, 44(3): 222-229. |
| 61 | ALVES N M, MANO J F, BALAGUER E, et al. Glass transition and structural relaxation in semi-crystalline poly(ethylene terephthalate): a DSC study[J]. Polymer, 2002, 43(15): 4111-4122. |
| 62 | TOURNIER V, TOPHAM C M, GILLES A, et al. An engineered PET depolymerase to break down and recycle plastic bottles[J]. Nature, 2020, 580(7802): 216-219. |
| 63 | WEI Ren, OESER T, BARTH M, et al. Turbidimetric analysis of the enzymatic hydrolysis of polyethylene terephthalate nanoparticles[J]. Journal of Molecular Catalysis B Enzymatic, 2014, 103: 72-78. |
| 64 | RIBITSCH D, ACERO E H, PRZYLUCKA A, et al. Enhanced cutinase-catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins[J]. Applied and Environmental Microbiology, 2015, 81(11): 3586-3592. |
| 65 | RIBITSCH D, YEBRA AO, ZITZENBACHER S, et al. Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing PET hydrolysis[J]. Biomacromolecules, 2013, 14(6): 1769–177. |
| 66 | GAMERITH C, ACERO E H, PELLIS A, et al. Improving enzymatic polyurethane hydrolysis by tuning enzyme sorption[J]. Polymer Degradation and Stability, 2016, 132: 69-77. |
| 67 | SILVA C, Shi DA, SILVA N, et al. Engineered Thermobifida fusca cutinase with increased activity on polyester substrates[J]. Biotechnology Journal, 2011, 6(10): 1230-1239. |
| 68 | WEI Ren, OESER T, SCHMIDT J, et al. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition[J]. Biotechnology and Bioengineering, 2016, 113(8): 1658-1665. |
| 69 | CARNIEL A, VALONI É, NICOMEDES J, et al. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid[J]. Process Biochemistry, 2017, 59: 84-90. |
| 70 | BARTH M, HONAK A, OESER T, et al. A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films[J]. Biotechnology Journal, 2016, 11(8): 1082-1087. |
| 71 | PARKE D, GARCIA M A, ORNSTON L N. Cloning and genetic characterization of dca genes required for β-oxidation of straight-chain dicarboxylic acids in Acinetobacter sp. strain ADP1[J]. Applied and Environmental Microbiology, 2001, 67(10): 4817-4827. |
| 72 | CHOI Jun-Ho, KIM Tae-Kang, KIM Young-Mog, et al. Cloning and characterization of a gene cluster for cyclohexanone oxidation in Rhodococcus sp. TK6[J]. Journal of Microbiology & Biotechnology, 2006, 16(4): 511-518. |
| 73 | FRANDEN M A, JAYAKODY L N, LI Wing-Jin, et al. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization[J]. Metabolic Engineering, 2018, 48: 197-207. |
| 74 | MUCKSCHEL B, SIMON O, KLEBENSBERGER J, et al. Ethylene glycol metabolism by Pseudomonas putida [J]. Applied and Environmental Microbiology, 2012, 78(24): 8531-8539. |
| 75 | TRIFUNOVIĆ D, SCHUCHMANN K, MULLER V. Ethylene glycol metabolism in the acetogen Acetobacterium woodii [J]. Journal of Bacteriology, 2016, 198(7): 1058-1065. |
| 76 | LI Wing-Jin, NARANCIC T, KENNY S T, et al. Unraveling 1,4-butanediol metabolism in Pseudomonas putida KT2440[J]. Frontiers in Microbiology, 2020, 11: 382. |
| 77 | SASOH M, MASAI E, ISHIBASHI S, et al. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6[J]. Applied and Environmental Microbiology, 2006, 72(3): 1825-1832. |
| 78 | SHIGEMATSU T, YUMIHARA K, UEDA Y, et al. Purification and gene cloning of the oxygenase component of the terephthalate 1,2-dioxygenase system from Delftia tsuruhatensis strain T7[J]. FEMS Microbiology Letters, 2003, 220(2): 255-260. |
| 79 | CHOI Ki Young, KIM Dockyu, Woo Jun SUL, et al. Molecular and biochemical analysis of phthalate and terephthalate degradation by Rhodococcussp. strain DK17[J]. FEMS Microbiology Letters, 2005, 252(2): 207-213. |
| 80 | KIM Hee Taek, KIM Jae Kyun, Hyun Gil CHA, et al. Biological valorization of poly(ethylene terephthalate) monomers for upcycling waste PET[J]. ACS Sustainable Chemistry & Engineering, 2019, 7(24): 19396-19406. |
| 81 | KENNY S T, RUNIC J N, KAMINSKY W, et al. Development of a bioprocess to convert PET derived terephthalic acid and biodiesel derived glycerol to medium chain length polyhydroxyalkanoate[J]. Applied Microbiology and Biotechnology, 2012, 95(3): 623-633. |
| 82 | ESPINOSA M J C, BLANCO A C, SCHMIDGALL T, et al. Toward biorecycling: isolation of a soil bacterium that grows on a polyurethane oligomer and monomer[J]. Frontiers in Microbiology, 2020, 11: 404. |
| 83 | TENAGY, PARK Jun Seok, IWAMA R, et al. Involvement of acyl-CoA synthetase genes in n-alkane assimilation and fatty acid utilization in yeast Yarrowia lipolytica [J]. FEMS Yeast Research, 2015, 15(4): fov031. |
| 84 | PATRAUCHAN M A, FLORIZONE C, EAPEN S, et al. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1[J]. Journal of Bacteriology, 2008, 190(1): 37-47. |
| 85 | PARK Mi-So, Jong-Won BAE, HAN Ju-Hee, et al. Characterization of styrene catabolic genes of Pseudomonas putida SN1 and construction of a recombinant Escherichia coli containing styrene monooxygenase gene for the production of (S)-styrene oxide[J]. Journal of Microbiology & Biotechnology, 2006, 16(7): 1032-1040. |
| 86 | ITOH N, YOSHIDA K, OKADA K. Isolation and identification of styrene-degrading Corynebacterium strains, and their styrene metabolism[J]. Bioscience, Biotechnology and Biochemistry, 1996, 60(11): 1826-1830. |
| 87 | TODA H, ITOH N. Isolation and characterization of styrene metabolism genes from styrene-assimilating soil bacteria Rhodococcus sp. ST-5 and ST-10[J]. Journal of Bioscience & Bioengineering, 2012, 113(1): 12-19. |
| 88 | OELSCHLÄGEL M, ZIMMERLING J, TISCHLER D. A review: the styrene metabolizing cascade of side-chain oxygenation as biotechnological basis to gain various valuable compounds[J]. Frontiers in Microbiology, 2018, 9: 490. |
| 89 | NIKODINOVIC-RUNIC J, CASEY E, DUANE G F, et al. Process analysis of the conversion of styrene to biomass and medium chain length polyhydroxyalkanoate in a two-phase bioreactor[J]. Biotechnology and Bioengineering, 2011, 108(10): 2447-2455. |
| 90 | SEN S K, RAUT S. Microbial degradation of low density polyethylene (LDPE): a review[J]. Journal of Environmental Chemical Engineering, 2015, 3(1): 462-473. |
| 91 | KENNY S T, RUNIC J N, KAMINSKY W, et al. Up-cycling of PET (polyethylene terephthalate) to the biodegradable plastic PHA (polyhydroxyalkanoate)[J]. Environmental Science & Technology, 2008, 42(20): 7696-7701. |
| 92 | CHRISTOVA N, TULEVA B, LALCHEV Z, et al. Rhamnolipid biosurfactants produced by Renibacterium salmoninarum 27BN during growth on n-hexadecane[J]. Ztschrift Für Naturforschung C, 2004, 59(1/2): 70-74. |
| 93 | ABDEL-MAWGOUD A M, LEPINE F, DEZIEL E. A stereospecific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants[J]. Chemistry & Biology, 2014, 21(1): 156-164. |
| 94 | MIHRETEAB M, STUBBLEFIELD B A, GILBERT E S. Microbial bioconversion of thermally depolymerized polypropylene by Yarrowia lipolytica for fatty acid production[J]. Applied Microbiology and Biotechnology, 2019, 103(18): 7729-7740. |
| 95 | CATUR UTOMO RN, LI Wing-Jin, TISO T, et al. Defined microbial mixed culture for utilization of polyurethane monomers[J]. ACS Sustainable Chemistry & Engineering, 2020, 8(47): 17466-17474. |
| 96 | PALM G J, REISKY L, BÖTTCHER D, et al. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate[J]. Nature Communications, 2019, 10(1): 1717. |
| 97 | AUSTIN H P, ALLEN M D, DONOHOE B S, et al. Characterization and engineering of a plastic-degrading aromatic polyesterase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 155: E4350-E4357. |
| 98 | INDERTHAL H, TAI Siew Leng, HARRISON S T L. Non-hydrolyzable plastics - an interdisciplinary look at plastic bio-oxidation[J]. Trends in Biotechnology, 2021, 39(1): 12-23. |
| 99 | YANG Shanshan, BRANDON A M, FLANAGAN J C A, et al. Biodegradation of polystyrene wastes in yellow mealworms (larvae of Tenebrio molitor Linnaeus): factors affecting biodegradation rates and the ability of polystyrene-fed larvae to complete their life cycle[J]. Chemosphere, 2018, 191: 979-989. |
| 100 | BRANDON A M, GAO Shuhong, TIAN Renmao, et al. Biodegradation of polyethylene and plastic mixtures in mealworms (larvae of Tenebrio molitor) and effects on the gut microbiome[J]. Environmental Science & Technology, 2018, 52(11): 6526-6533. |
| 101 | PENG Boyu, SU Yiming, CHEN Zhibin, et al. Biodegradation of polystyrene by dark (Tenebrio obscurus) and yellow (Tenebrio molitor) mealworms (Coleoptera: Tenebrionidae)[J]. Environmental Science & Technology, 2019, 53(9): 5256-5265. |
| 102 | KONG Hyun Gi, KIM Hyun Ho, CHUNG Joon-hui, et al. The Galleria mellonella hologenome supports microbiota-independent metabolism of long-chain hydrocarbon beeswax[J]. Cell Reports, 2019, 26(9): 2451-2464.e5. |
| 103 | JEON Hyun Jeong, KIM Mal Nam. Comparison of the functional characterization between alkane monooxygenases for low-molecular-weight polyethylene biodegradation[J]. International Biodeterioration & Biodegradation, 2016, 114: 202-208. |
| 104 | GUAN Zhengbing, LUO Quan, WANG Haoran, et al. Bacterial laccases: promising biological green tools for industrial applications[J]. Cellular and Molecular Life Sciences, 2018, 75: 3569-3592. |
| 105 | SHIRKE A N, WHITE C, ENGLAENDER J A, et al. Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: mechanism and effect on PET hydrolysis[J]. Biochemistry, 2018, 57(7): 1190-1200. |
| 106 | NICOUD L, OWCZARZ M, AROSIO P, et al. A multiscale view of therapeutic protein aggregation: a colloid science perspective[J]. Biotechnology Journal, 2015, 10(3): 367-378. |
| 107 | MAJHI P R, GANTA R R, VANAM R P, et al. Electrostatically driven protein aggregation: β-lactoglobulin at low ionic strength[J]. Langmuir, 2006, 22(22): 9150-9159. |
| 108 | JIANG Liguo, CAO Siqin, Pak-Hang CHEUNG P, et al. Real-time monitoring of hydrophobic aggregation reveals a critical role of cooperativity in hydrophobic effect[J]. Nature Communications, 2017, 8: 15639. |
| 109 | KAMAL M Z, AHMAD S, MOLUGU T R, et al. In vitro evolved non-aggregating and thermostable lipase: structural and thermodynamic investigation[J]. Journal of Molecular Biology, 2011, 413(3): 726-741. |
| 110 | MATSUI D, NAKANO S, DADASHIPOUR M, et al. Rational identification of aggregation hotspots based on secondary structure and amino acid hydrophobicity[J]. Scientific Reports, 2017, 7(1): 9558. |
| 111 | SHENTAL-BECHOR D, LEVY Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization [J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(24): 8256-8261. |
| 112 | CHEN Zhi, ZHAO Wenqi, XING Ruizhi, et al. Enhanced in situ biodegradation of microplastics in sewage sludge using hyperthermophilic composting technology[J]. Journal of Hazardous Materials, 2019, 384: 121271. |
| 113 | SOLOMON K V, HAITJEMA C H, HENSKE J K, et al. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes[J]. Science, 2016, 351(6278): 1192-1195. |
| 1 | PLASTICSEUROPE. Plastics—the Facts 2020. An analysis of European plastics production, demand and waste data[R]. Brussels: Plastics Europe, Association of Plastic Manufacturers, 2020. |
| 2 | AMIRKHANIAN S. Utilization of scrap plastics in asphalt binders[M]// PACHECO-TORGAL F, AMIRKHANIAN S, WANG Hao, et al. Eco-efficient pavement construction materials. Sawston, Cambridge: Woodhead Publishing, 2020. |
| 3 | KAZA S, YAO L C, BHADA-TATA P, et al. What a waste 2.0: a global snapshot of solid waste management to 2050[M]. Washington: World Bank Group, 2018. |
| 4 | 中国物资再生协会再生塑料分会.中国再生塑料行业发展报告2019—2020 [R]. 北京:中国物资再生协会再生塑料分会, 2020.China National Resources Recycling Association. 2019—2020 Development report of China plastic recycling industry[R]. Beijing: China National Resources Recycling Association, 2020. |
| 5 | JAMBECK J R, GEYER R, WILCOX C, et al. Plastic waste inputs from land into the ocean[J]. Science, 2015, 347(6223): 768-771. |
| 6 | LAMBERT S, WAGNER M. Characterisation of nanoplastics during the degradation of polystyrene[J]. Chemosphere, 2016, 145: 265-268. |
| 7 | 魏鑫嘉, 刘博洋, 王鸣, 等. 废塑料裂解及塑料油精制研究进展[J]. 工业催化, 2019, 27(2): 31-34. |
| WEI Xinjia, LIU Boyang, WANG Ming, et al. Progress inpyrolysis of waste plastics and refining of waste plastic oil[J]. Industrial Catalysis, 2019, 27(2): 31-34. | |
| 8 | GEYER R, JAMBECK J R, LAW K L. Production, use, and fate of all plastics ever made[J]. Science Advances, 2017, 3(7): e1700782. |
| 9 | IWATA T. Biodegradable and bio-based polymers: future prospects of eco-friendly plastics[J]. Angewandte Chemie International Edition, 2015, 54(11): 3210-3215. |
| 10 | SIVAN A. New perspectives in plastic biodegradation[J]. Current Opinion in Biotechnology, 2011, 22(3): 422-426. |
| 11 | ARAÚJO R, SILVA C, O'NEILL A, et al. Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers[J]. Journal of Biotechnology, 2007, 128(4): 849-857. |
| 12 | NIMCHUA T, EVELEIGH DE, SANGWATANAROJ U, et al. Screening of tropical fungi producing polyethylene terephthalate-hydrolyzing enzyme for fabric modification[J]. Journal of Industrial Microbiology & Biotechnology, 2008, 35(8): 843-850. |
| 13 | RONKVIST S Å, XIE Wenchun, LU Wenhua, et al. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate)[J]. Macromolecules, 2009, 42(14): 5128-5138. |
| 14 | KLEEBERG I, HETZ C, KROPPENSTEDT R M, et al. Biodegradation of aliphatic-aromatic copolyesters by Thermomonospora fusca and other thermophilic compost isolates[J]. Applied & Environmental Microbiology, 1998, 64(5): 1731-1735. |
| 15 | R-J MÜLLER, SCHRADER H, PROFE J, et al. Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca [J]. Macromolecular Rapid Communications, 2010, 26(17): 1400-1405. |
| 16 | THEN J, WEI Ren, OESER T, et al. A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate[J]. FEBS Open Bio, 2016, 6(5): 425-432. |
| 17 | THEN J, WEI Ren, OESER T, et al. Ca2+and Mg2+binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca [J]. Biotechnology Journal, 2015, 10(4): 592-598. |
| 18 | HU Xiaoping, OSAKI S, HAYASHI M, et al. Degradation of a terephthalate-containing polyester by Thermophilic Actinomycetes and Bacillus Species derived from composts[J]. Journal of Polymers and the Environment, 2008, 16(4): 103-108. |
| 19 | YOSHIDA S, HIRAGA K, TAKEHANA T, et al. A bacterium that degrades and assimilates poly(ethylene terephthalate)[J]. Science, 2016, 351(6278): 1196-1199. |
| 20 | MATHUR G, PRASAD R. Degradation of polyurethane by Aspergillus flavus (ITCC 6051) isolated from soil[J]. Applied Biochemistry & Biotechnology, 2012, 167(6): 1595-1602. |
| 21 | KHAN S, NADIR S, SHAH Z U, et al. Biodegradation of polyester polyurethane by Aspergillus tubingensis [J]. Environmental Pollution, 2017, 225(1): 469-480. |
| 22 | OSMAN M, SATTI SM, LUQMAN A, et al. Degradation of polyester polyurethane by Aspergillus sp. strain S45 isolated from soil[J]. Journal of Polymers and the Environment, 2018, 26(1): 301-310. |
| 23 | ÁLVAREZ-BARRAGÁN J, DOMÍNGUEZ-MALFAVÓN L, VARGAS-SUÁREZ M, et al. Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams[J]. Applied and Environmental Microbiology, 2016, 82(17): 5225-5235. |
| 114 | CHEN Chunchi, DAI Longhai, MA Lixin, et al. Enzymatic degradation of plant biomass and synthetic polymers[J]. Nature Reviews Chemistry, 2020, 4(3): 114-126. |
| 115 | ESCAPA I F, GARCÍA J L, BÜHLER B, et al. The polyhydroxyalkanoate metabolism controls carbon and energy spillage in Pseudomonas putida [J]. Environmental Microbiology, 2012, 14(4): 1049-1063. |
| 116 | WIERCKX N, PRIETO M A, POMPOSIELLO P, et al. Plastic waste as a novel substrate for industrial biotechnology[J]. Microbial Biotechnology, 2015, 8(6): 900-903. |
| 117 | RU Jiakang, HUO Yixin, YANG Yu. Microbial degradation and valorization of plastic wastes[J]. Frontiers in Microbiology, 2020, 11: 442. |
| 24 | WEBB H K, ARNOTT J, CRAWFORD R J, et al. Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate)[J]. Polymers, 2012, 5(1): 1-18. |
| 25 | MÜELLER R J. Biological degradation of synthetic polyesters — enzymes as potential catalysts for polyester recycling [J]. Process Biochemistry, 2006, 41(10): 2124-2128. |
| 26 | 李江华, 刘龙, 陈晟, 等. 角质酶的研究进展[J]. 生物工程学报, 2009, 25(12): 1829-1837. |
| LI Jianghua, LIU Long, CHEN Sheng, et al. Advances in cutinase research[J]. Chinese Journal of Biotechnology, 2009, 25(12): 1829-1837. | |
| 27 | KLEEBERG I, WELZEL K, VANDENHEUVEL J, et al. Deckwer. Characterization of a new extracellular hydrolase from Thermobifida fusca degrading aliphatic-aromatic copolyesters [J]. Biomacromolecules, 2005, 6: 262-270. |
| 28 | FEDER D. Humicola insolens cutinase; a novel catalyst for polymer synthesis reactions[D]. New York: Polytechnic Institute of New York University, 2013. |
| 29 | SULAIMAN S, YOU Dong-Ju, KANAYA E, et al. Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase[J]. Biochemistry, 2014, 53(11): 1858-1869. |
| 30 | CHEN Sheng, SU Lingqia, BILLIG S, et al. Biochemical characterization of the cutinases from Thermobifida fusca [J]. Journal of Molecular Catalysis B Enzymatic, 2010, 63(3/4): 121-127. |
| 31 | ACERO E H, RIBITSCH D, STEINKELLNER G, et al. Enzymatic surface hydrolysis of PET: effect of structural diversity on kinetic properties of cutinases from Thermobifida [J]. Macromolecules, 2011, 44(12): 4632-4640. |
| 32 | AKUTSU Y, NAKAJIMA-KAMBE T, NOMURA N, et al. Purification and properties of a polyester polyurethane-degrading enzyme from Comamonas acidovorans TB-35[J]. Applied & Environmental Microbiology, 1998, 64(1): 62-67. |
| 33 | NOMURA N, SHIGENO-AKUTSU Y, NAKAJIMA-KAMBE T, et al. Cloning and sequence analysis of a polyurethane esterase of Comamonas acidovorans TB-35[J]. Journal of Fermentation & Bioengineering, 1998, 86(4): 339-345. |
| 34 | RUIZ C, HOWARD G T. Nucleotide sequencing of a polyurethanase gene (pulA) from Pseudomonas fluorescens [J]. International Biodeterioration & Biodegradation, 1999, 44(2/3): 127-131. |
| 35 | STERN R V, HOWARD G T. The polyester polyurethanase gene (pueA) from Pseudomonas chlororaphis encodes a lipase[J]. FEMS Microbiology Letters, 2000, 185(2): 163–168. |
| 36 | HOWARD G T, CROTHER B, VICKNAIR J. Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas chlororaphis [J]. International Biodeterioration & Biodegradation, 2001, 47(3): 141-149. |
| 37 | SCHMIDT J, WEI Ren, OESER T, et al. Degradation of polyester polyurethane by bacterial polyester hydrolases[J]. Polymers, 2017, 9(2): 65. |
| 38 | MAHAJAN N, GUPTA P. New insights into the microbial degradation of polyurethanes[J]. RSC Advances, 2015, 5(52): 41839-41854. |
| 39 | MAGNIN A, POLLET E, PHALIP V, et al. Evaluation of biological degradation of polyurethanes[J]. Biotechnology Advances, 2020, 39: 107457. |
| 40 | 彭瑞婷, 夏孟丽, 茹家康, 等. 聚氨酯塑料的微生物降解[J]. 生物工程学报, 2018, 34(9): 1398-1409. |
| PENG Ruiting, XIA Mengli, RU Jiakang, et al. Microbial degradation of polyurethane plastics[J]. Chinese Journal of Biotechnology, 2018, 34(9): 1398-1409. | |
| 41 | PHUA S K, CASTILLO E, ANDERSON J M, et al. Biodegradation of a polyurethane in vitro[J]. Journal of Biomedical Materials Research Part A, 2010, 21(2): 231-46. |
| 42 | CAMPIÑEZ M D, AGUILAR-DE-LEYVA Á, FERRIS C, et al. Study of the properties of the new biodegradable polyurethane PU (TEG-HMDI) as matrix forming excipient for controlled drug delivery[J]. Drug Development and Industrial Pharmacy, 2013, 39(11): 1758-1764. |
| 43 | 许楹, 殷超凡, 岳纹龙, 等. 石油基塑料的微生物降解[J]. 生物工程学报, 2019, 35(11): 2092-2103. |
| XU Ying, YIN Chaofan, YUE Wenlong, et al. Microbial degradation of petroleum-based plastics[J]. Chinese Journal of Biotechnology, 2019, 35(11): 2092-2103. | |
| 44 | 韩秋霞, 王庆昭, 张萌. 改性PE膜的生物可降解性研究[J]. 塑料工业, 2009, 37(10): 48-51. |
| HAN Qiuxia, WANG Qingzhao, ZHANG Meng. Study on biodegradability of modified PE film[J]. China Plastics Industry, 2009, 37(10): 48-51. | |
| 45 | BALASUBRAMANIAN V, NATARAJAN K, HEMAMBIKA B, et al. High-density polyethylene (HDPE)-degrading potential bacteria from marine ecosystem of Gulf of Mannar, India[J]. Letters in Applied Microbiology, 2010, 51(2): 205-211. |
| 46 | YANG Yu, YANG Jun, WU Weimin, et al. Biodegradation and mineralization of polystyrene by plastic-eating mealworms (I): Chemical and physical characterization and isotopic tests[J]. Environmental Science & Technology, 2015, 49(20): 12080-12086. |
| 47 | YANG Yu, YANG Jun, WU Weimin, et al. Biodegradation and mineralization of polystyrene by plastic-eating mealworms (Ⅱ): Role of gut microorganisms[J]. Environmental Science & Technology, 2015, 49(20): 12087-12093. |
| 48 | TIAN Lili, KOLVENBACH Boris, CORVINI Nora, et al. Mineralisation of 14C-labelled polystyrene plastics by Penicillium variabile after ozonation pre-treatment[J]. New Biotechnology, 2017, 38: 101-105. |
| 49 | Tribedi P, Sil A K. Low-density polyethylene degradation by Pseudomonas sp. AKS2 biofilm.Environmental Science and Pollution Research, 2013, 20(6): 4146–4153. |
| 50 | Gilan I, Hadar Y, Sivan A. Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber [J]. Applied Microbiology and Biotechnology, 2004, 65(1): 97-104. |
| 51 | OIKAWA E, LINN KT, ENDO T, et al. Isolation and characterization of polystyrene degrading microorganisms for zero emission treatment of expanded polystyrene[J]. Environmental Engineering Research, 2003, 40: 373-379. |
| 52 | YANG Yu, YANG Jun, WU Weimin, et al. Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms[J]. Environmental Science & Technology, 2014, 48(23): 13776-13784. |
| [1] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [2] | LIU Weisong, ZHANG Kuncheng, CUI Huijuan, ZHU Zhiguang, ZHANG Yiheng, ZHANG Lingling. Electro-assisted carbon dioxide biotransformation [J]. Synthetic Biology Journal, 2023, 4(6): 1191-1222. |
| [3] | MING Yang, CHEN Bin, HUANG Xiaoqiang. Recent advances in photoenzymatic synthesis [J]. Synthetic Biology Journal, 2023, 4(4): 651-675. |
| [4] | GUO Shuyuan, WU Lianghuan, LIU Xiangjian, WANG Bo, YU Tao. Developing C1-based metabolic network in methylotrophy for biotransformation [J]. Synthetic Biology Journal, 2022, 3(1): 116-137. |
| [5] | XIONG Liangbin, SONG Lu, ZHAO Yunqiu, LIU Kun, LIU Yongjun, WANG Fengqing, WEI Dongzhi. Green biomanufacturing of steroids: from biotransformation to de novo synthesis by microorganisms [J]. Synthetic Biology Journal, 2021, 2(6): 942-963. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
