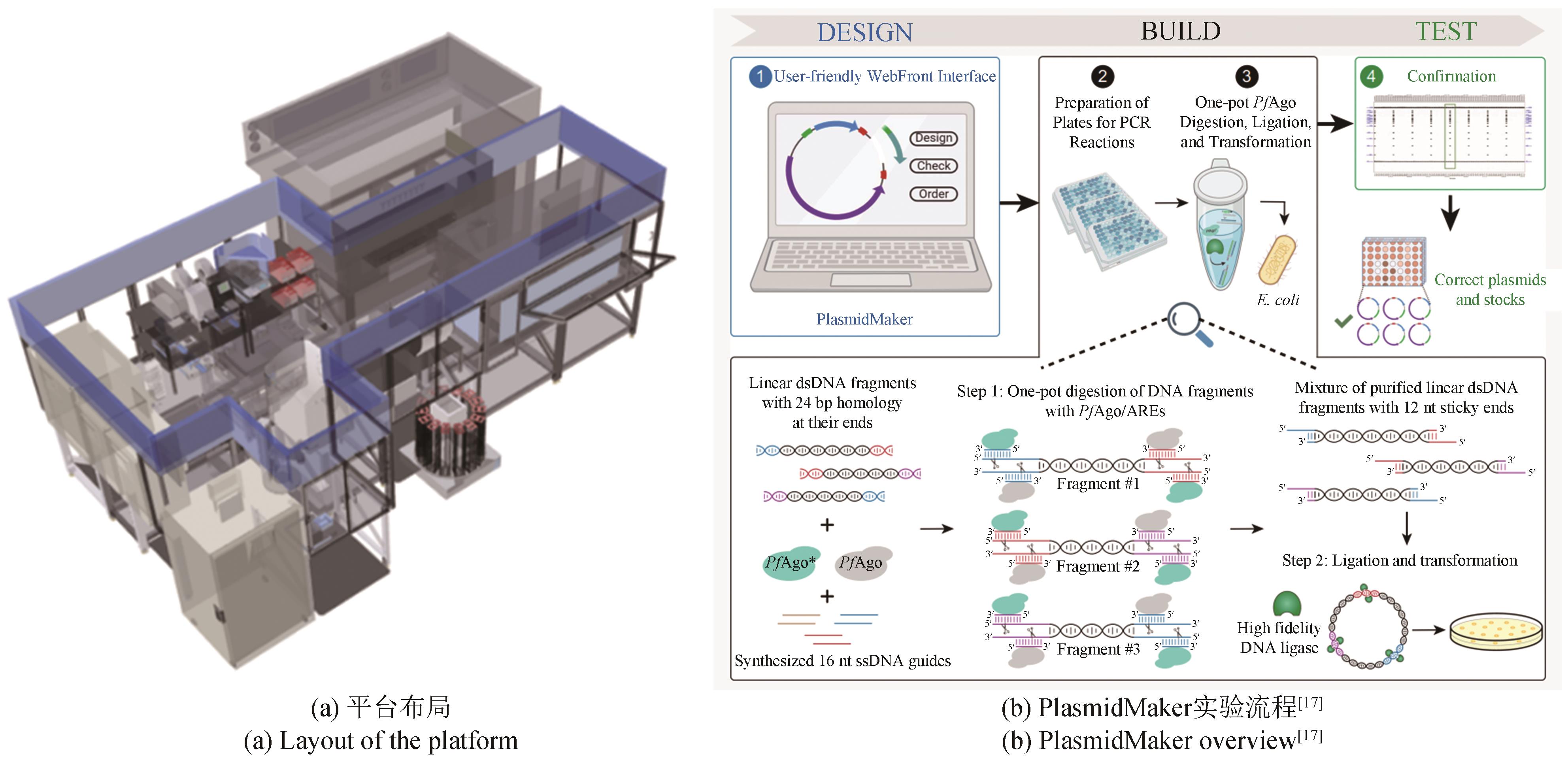
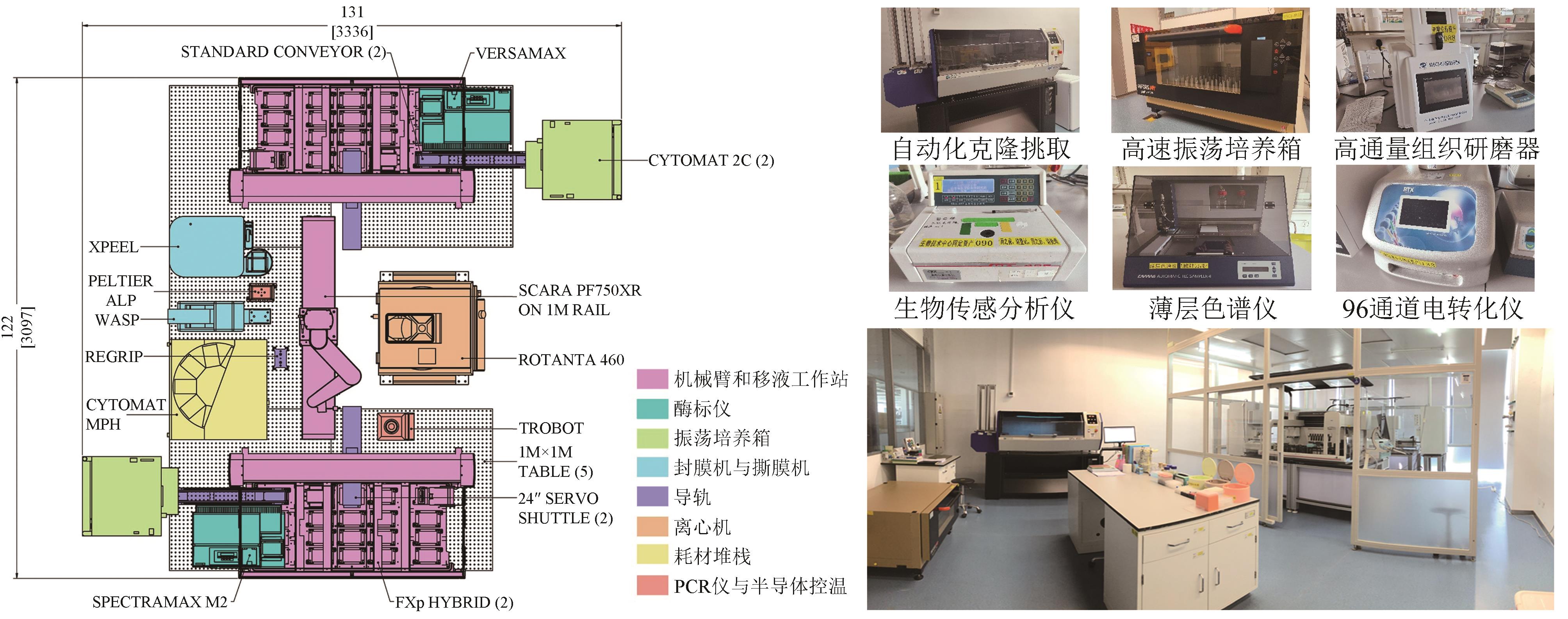
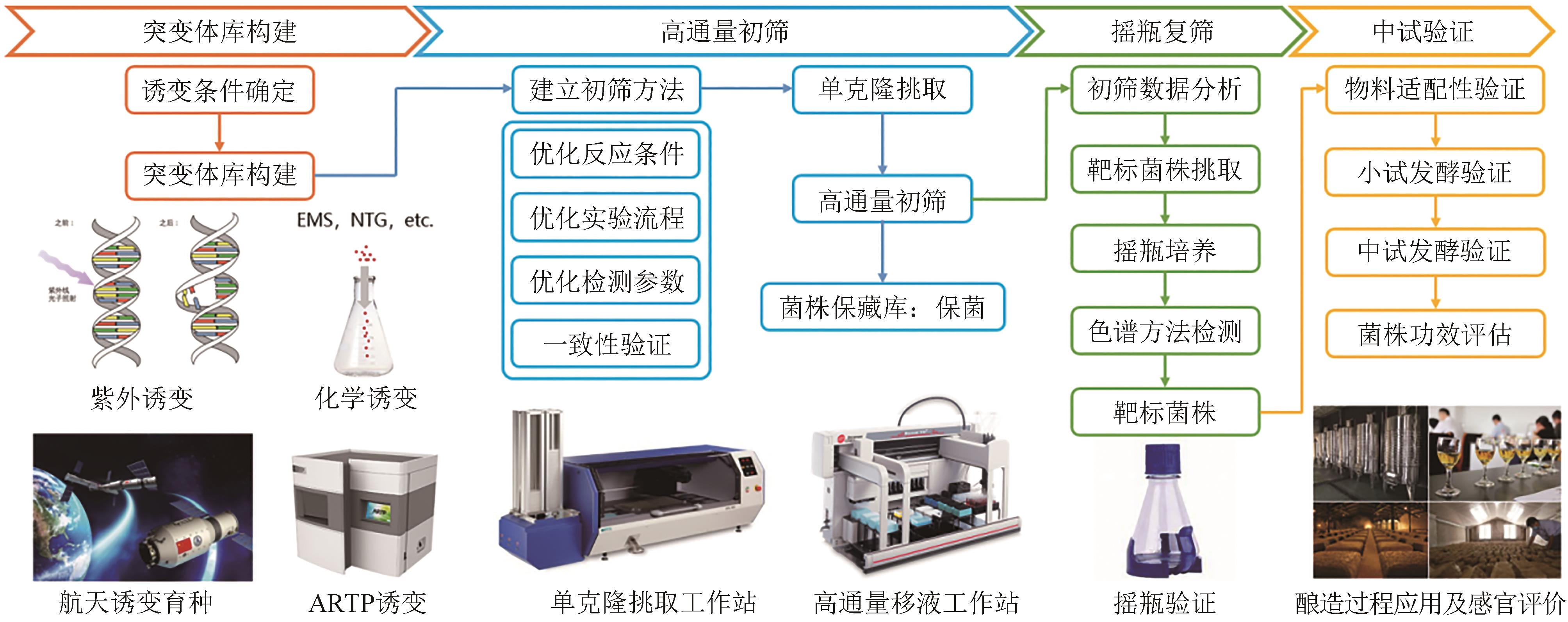
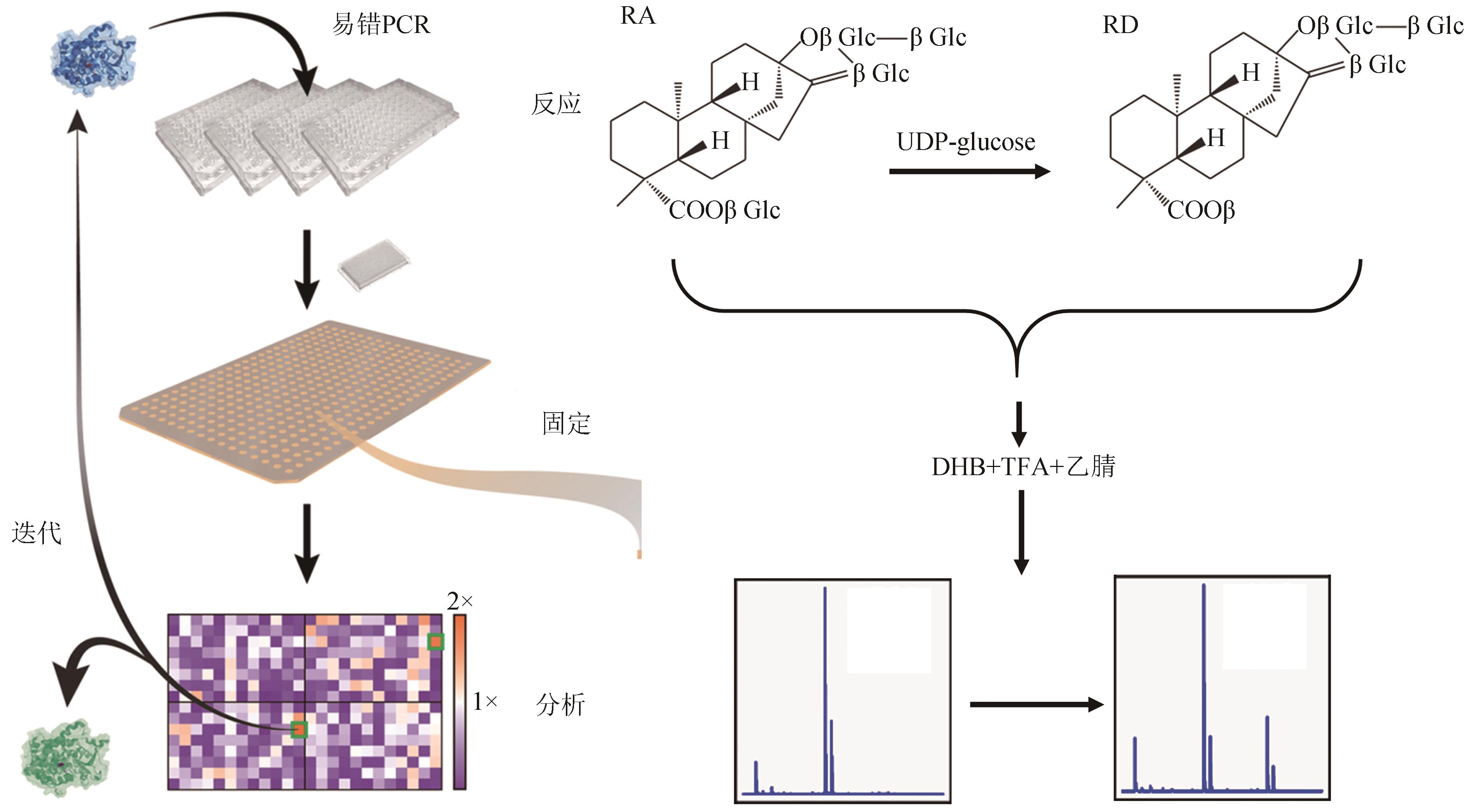
Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (5): 892-903.DOI: 10.12211/2096-8280.2023-024
• Invited Review • Previous Articles Next Articles
Biofoundry and its industrial application
ZHAO Guomiao1,2, YANG Xin1,2, ZHANG Yuan1,2, WANG Jing1,2, TAN Jian1,2, WEI Chao1,2, ZHOU Nana1,2, LI Fan1,2, WANG Xiaoyan1,2
- 1.Nutrition & Health Research Institute,COFCO Corporation,Beijing 102209,China
2.Beijing Key Laboratory of Nutrition,Health and Food Safety,Beijing 102209,China
-
Received:2023-03-17Revised:2023-05-08Online:2023-11-15Published:2023-10-31 -
Contact:WANG Xiaoyan
生物设施平台及其工业应用
赵国淼1,2, 杨鑫1,2, 张媛1,2, 王靖1,2, 谭剑1,2, 魏超1,2, 周娜娜1,2, 李凡1,2, 王小艳1,2
- 1.中粮营养健康研究院有限公司,北京 102209
2.营养健康与食品安全北京市重点实验室,北京 102209
-
通讯作者:王小艳 -
作者简介:赵国淼 (1989—),男,博士,工程师。研究方向为工业微生物改造与高通量筛选、计算生物学与生物信息学。E-mail:zhaoguomiao@cofco.com王小艳 (1980—),女,博士,正高级工程师。研究方向围绕淀粉质原料生物加工过程工业菌株和酶制剂的开发,主要聚焦生物燃料乙醇、生物基材料、功能糖等领域。E-mail:wangxiaoyan@cofco.com -
基金资助:国家重点研发计划“合成生物学”重点专项,合成生物学自动化铸造平台关键技术研发(2018YFA0902900)
CLC Number:
Cite this article
ZHAO Guomiao, YANG Xin, ZHANG Yuan, WANG Jing, TAN Jian, WEI Chao, ZHOU Nana, LI Fan, WANG Xiaoyan. Biofoundry and its industrial application[J]. Synthetic Biology Journal, 2023, 4(5): 892-903.
赵国淼, 杨鑫, 张媛, 王靖, 谭剑, 魏超, 周娜娜, 李凡, 王小艳. 生物设施平台及其工业应用[J]. 合成生物学, 2023, 4(5): 892-903.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-024
| 筛选方法 | 检测信号 | 灵敏度 | 通量 |
|---|---|---|---|
| MTP[ | 吸光度和荧光强度 | 一般 | 106个/天 |
| FACS[ | 荧光强度 | 高 | 108个/时 |
| DMF[ | 荧光强度、拉曼光谱、吸光度、质谱 | 高 | 108个/天 |
Table 1 Comparison of three screening methods
| 筛选方法 | 检测信号 | 灵敏度 | 通量 |
|---|---|---|---|
| MTP[ | 吸光度和荧光强度 | 一般 | 106个/天 |
| FACS[ | 荧光强度 | 高 | 108个/时 |
| DMF[ | 荧光强度、拉曼光谱、吸光度、质谱 | 高 | 108个/天 |
| 1 | LEE S Y, KIM H U, CHAE T U, et al. A comprehensive metabolic map for production of bio-based chemicals[J]. Nature Catalysis, 2019, 2(1): 18-33. |
| 2 | CHEN X L, GAO C, GUO L, et al. DCEO biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals[J]. Chemical Reviews, 2018, 118(1): 4-72. |
| 3 | RUGBJERG P, SOMMER M O A. Overcoming genetic heterogeneity in industrial fermentations[J]. Nature Biotechnology, 2019, 37(8): 869-876. |
| 4 | WEHRS M, TANJORE D, ENG T, et al. Engineering robust production microbes for large-scale cultivation[J]. Trends in Microbiology, 2019, 27(6): 524-537. |
| 5 | LEE S Y, KIM H U. Systems strategies for developing industrial microbial strains[J]. Nature Biotechnology, 2015, 33(10): 1061-1072. |
| 6 | ZHANG X, ZHANG X M, XU G Q, et al. Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve L-serine yield in Corynebacterium glutamicum [J]. Applied Microbiology and Biotechnology, 2018, 102(14): 5939-5951. |
| 7 | GU Y, XU X H, WU Y K, et al. Advances and prospects of Bacillus subtilis cellular factories: from rational design to industrial applications[J]. Metabolic Engineering, 2018, 50: 109-121. |
| 8 | CARBONELL P, JERVIS A J, ROBINSON C J, et al. An automated Design-Build-Test-Learn pipeline for enhanced microbial production of fine chemicals[J]. Communications Biology, 2018, 1: 66. |
| 9 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 10 | QUAGLIA D, EBERT M C C J C, MUGFORD P F, et al. Enzyme engineering: a synthetic biology approach for more effective library generation and automated high-throughput screening[J]. PLoS One, 2017, 12(2): e0171741. |
| 11 | ZHANG Y V, ROCKWOOD A. Impact of automation on mass spectrometry[J]. Clinica Chimica Acta, 2015, 450: 298-303. |
| 12 | LONGWELL C K, LABANIEH L, COCHRAN J R. High-throughput screening technologies for enzyme engineering[J]. Current Opinion in Biotechnology, 2017, 48: 196-202. |
| 13 | RAN C, MISHRA S, TONG S, et al. Engineering biological systems using automated biofoundries[J]. Metabolic Engineering, 2017, 42: 98-108. |
| 14 | ZHANG J Z, CHEN Y C, FU L H, et al. Accelerating strain engineering in biofuel research via build and test automation of synthetic biology[J]. Current Opinion in Biotechnology, 2021, 67: 88-98. |
| 15 | LE FEUVRE R A, SCRUTTON N S. A living foundry for Synthetic Biological Materials: a synthetic biology roadmap to new advanced materials[J]. Synthetic & Systems Biotechnology, 2018, 3(2): 105-112. |
| 16 | HILLSON N, CADDICK M, CAI Y Z, et al. Building a global alliance of biofoundries[J]. Nature Communications, 2019, 10: 2040. |
| 17 | ENGHIAD B, XUE P, SINGH N, et al. PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction[J]. Nature Communications, 2022, 13: 2697. |
| 18 | ZHAO H M. Illinois biological foundry for advanced biomanufacturing (iBioFAB)[C/OL]. Synthetic Biology: Engineering, Evolution, and Design Conference 2015, SEED 2015, 2015, 2: 784-785[2023-03-01]. . |
| 19 | HAMEDIRAD M, CHAO R, WEISBERG S, et al. Towards a fully automated algorithm driven platform for biosystems design[J]. Nature Communications, 2019, 10: 5150. |
| 20 | XU K, QIN L, BAI W X, et al. Multilevel defense system (MDS) relieves multiple stresses for economically boosting ethanol production of industrial Saccharomyces cerevisiae [J]. ACS Energy Letters, 2020, 5(2): 572-582. |
| 21 | LIU W S, JIANG R R. Combinatorial and high-throughput screening approaches for strain engineering[J]. Applied Microbiology and Biotechnology, 2015, 99(5): 2093-2104. |
| 22 | ALPER H, MIYAOKU K, STEPHANOPOULOS G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets[J]. Nature Biotechnology, 2005, 23(5): 612-616. |
| 23 | ÖZAYDIN B, BURD H, LEE T S, et al. Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production[J]. Metabolic Engineering, 2013, 15: 174-183. |
| 24 | ZELCBUCH L, ANTONOVSKY N, BAR-EVEN A, et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics[J]. Nucleic Acids Research, 2013, 41(9): e98. |
| 25 | LEE J H, LEE S H, YIM S S, et al. Quantified high-throughput screening of Escherichia coli producing poly(3-hydroxybutyrate) based on FACS[J]. Applied Biochemistry and Biotechnology, 2013, 170(7): 1767-1779. |
| 26 | TYO K E J, JIN Y S, ESPINOZA F A, et al. Identification of gene disruptions for increased poly-3-hydroxybutyrate accumulation in Synechocystis PCC 6803[J]. Biotechnology Progress, 2009, 25(5): 1236-1243. |
| 27 | KLEIN-MARCUSCHAMER D, SANTOS C N S, YU H M, et al. Mutagenesis of the bacterial RNA polymerase alpha subunit for improvement of complex phenotypes[J]. Applied and Environmental Microbiology, 2009, 75(9): 2705-2711. |
| 28 | ALPER H, MOXLEY J, NEVOIGT E, et al. Engineering yeast transcription machinery for improved ethanol tolerance and production[J]. Science, 2006, 314(5805): 1565-1568. |
| 29 | BASAK S, GENG H F, JIANG R R. Rewiring global regulator cAMP receptor protein (CRP) to improve E. coli tolerance towards low pH[J]. Journal of Biotechnology, 2014, 173: 68-75. |
| 30 | LIU H M, YAN M, LAI C G, et al. gTME for improved xylose fermentation of Saccharomyces cerevisiae [J].Applied Biochemistry and Biotechnology, 2010, 160(2): 574-582. |
| 31 | CHONG H Q, HUANG L, YEOW J, et al. Improving ethanol tolerance of Escherichia coli by rewiring its global regulator cAMP receptor protein (CRP)[J]. PLoS One, 2013, 8(2): e57628. |
| 32 | HENNING H, LEGGEWIE C, POHL M, et al. Identification of novel benzoylformate decarboxylases by growth selection[J]. Applied and Environmental Microbiology, 2006, 72(12): 7510-7517. |
| 33 | PFLEGER B F, PITERA D J, SMOLKE C D, et al. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes[J]. Nature Biotechnology, 2006, 24(8): 1027-1032. |
| 34 | BOERSMA Y L, DRÖGE M J, VAN DER SLOOT A M, et al. A novel genetic selection system for improved enantioselectivity ofBacillus subtilis lipase A[J]. ChemBioChem, 2008, 9(7): 1110-1115. |
| 35 | BOLES E, OREB M. A growth-based screening system for hexose transporters in yeast[M/OL]//Methods in Molecular Biology. New York, NY: Springer New York, 2018, 1713: 123-135 [2023-03-01]. . |
| 36 | DIETRICH J A, MCKEE A E, KEASLING J D. High-throughput metabolic engineering: advances in small-molecule screening and selection[J]. Annual Review of Biochemistry, 2010, 79: 563-590. |
| 37 | LATCHMAN D S. Transcription factors: an overview[J]. The International Journal of Biochemistry & Cell Biology, 1997, 29(12): 1305-1312. |
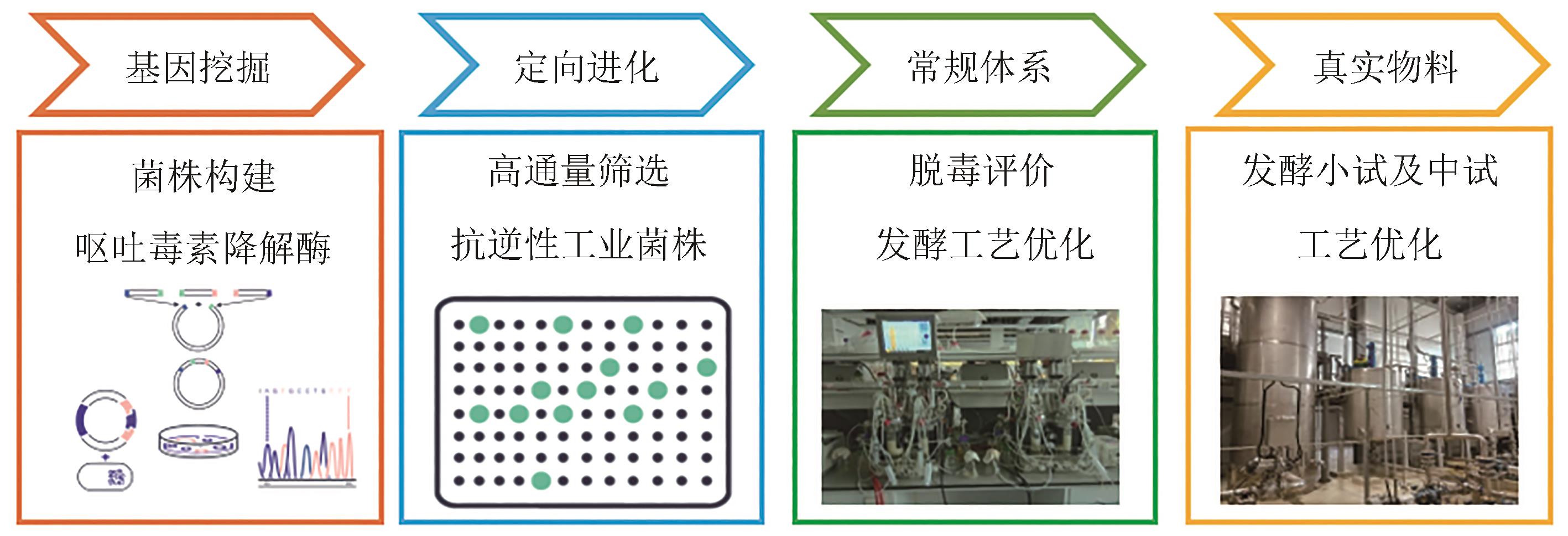
| 38 | BINDER S, SCHENDZIELORZ G, STÄBLER N, et al. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level[J]. Genome Biology, 2012, 13(5): R40. |
| 39 | MAHR R, GÄTGENS C, GÄTGENS J, et al. Biosensor-driven adaptive laboratory evolution of l-valine production in Corynebacterium glutamicum [J]. Metabolic Engineering, 2015, 32: 184-194. |
| 40 | BASTET L, TURCOTTE P, WADE J T, et al. Maestro of regulation: riboswitches orchestrate gene expression at the levels of translation, transcription and mRNA decay[J]. RNA Biology, 2018: 15(6): 679-682. |
| 41 | ECKDAHL T T, CAMPBELL A M, HEYER L J, et al. Programmed evolution for optimization of orthogonal metabolic output in bacteria[J]. PLoS One, 2015, 10(2): e0118322. |
| 42 | DIXON N, DUNCAN J N, GEERLINGS T, et al. Reengineering orthogonally selective riboswitches[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(7): 2830-2835. |
| 43 | WANG J L, WEI J H, SU S H, et al. Novel fluorescence resonance energy transfer optical sensors for vitamin B12 detection using thermally reduced carbon dots[J]. New Journal of Chemistry, 2015, 39(1): 501-507. |
| 44 | NGUYEN T T T, HUY B T, TAWFIK S M, et al. Highly selective and sensitive optosensing of glutathione based on fluorescence resonance energy transfer of upconversion nanoparticles coated with a Rhodamine B derivative[J]. Arabian Journal of Chemistry, 2020, 13(1): 2671-2679. |
| 45 | DING Y D, LI J, ENTERINA J R, et al. Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange[J]. Nature Methods, 2015, 12(3): 195-198. |
| 46 | FU X Z, ZHANG Y Y, XU Q, et al. Recent advances on sorting methods of high-throughput droplet-based microfluidics in enzyme directed evolution[J]. Frontiers in Chemistry, 2021, 9: 666867. |
| 47 | ZHANG Z D, GUO Q, WANG Y T, et al. High-throughput screening of microbial strains in large-scale microfluidic droplets[J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1105277. |
| 48 | UTHARALA R, GRAB A, VAFAIZADEH V, et al. A microfluidic Braille valve platform for on-demand production, combinatorial screening and sorting of chemically distinct droplets[J]. Nature Protocols, 2022, 17(12): 2920-2965. |
| 49 | KÖRFER G, PITZLER C, VOJCIC L, et al. In vitro flow cytometry-based screening platform for cellulase engineering[J]. Scientific Reports, 2016, 6: 26128. |
| 50 | WANG Y T, ZHANG X X, SHANG L R, et al. Thriving microfluidic technology[J]. Science Bulletin, 2021, 66(1): 9-12. |
| 51 | 涂然, 李世新, 李昊霓, 等. 液滴微流控技术在微生物工程菌株选育中的应用进展[J]. 合成生物学, 2023(1): 165-184. |
| TU R, LI S X, LI H N, et al. Advances and applications of droplet-based microfluidics in evolution and screening of engineered microbial strains[J]. Synthetic Biology Journal, 2023(1): 165-184. | |
| 52 | LEAMON J H, LINK D R, EGHOLM M, et al. Overview: methods and applications for droplet compartmentalization of biology[J]. Nature Methods, 2006, 3(7): 541-543. |
| 53 | GIELEN F, HOURS R, EMOND S, et al. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS)[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(47): E7383-E7389. |
| 54 | BARET J C, MILLER O J, TALY V, et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity[J]. Lab on a Chip, 2009, 9(13): 1850-1858. |
| 55 | GOTO H, KANAI Y, YOTSUI A, et al. Microfluidic screening system based on boron-doped diamond electrodes and dielectrophoretic sorting for directed evolution of NAD(P)-dependent oxidoreductases[J]. Lab on a Chip, 2020, 20(4): 852-861. |
| 56 | WANG X X, REN L H, SU Y T, et al. Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells[J]. Analytical Chemistry, 2017, 89(22): 12569-12577. |
| 57 | SESEN M, WHYTE G. Image-based single cell sorting automation in droplet microfluidics[J]. Scientific Reports, 2020, 10: 8736. |
| 58 | HOLLAND-MORITZ D A, WISMER M K, MANN B F, et al. Mass activated droplet sorting (MADS) enables high-throughput screening of enzymatic reactions at nanoliter scale[J]. Angewandte Chemie International Edition, 2020, 59(11): 4470-4477. |
| 59 | 王小艳, 秦磊, 刘辉, 等. 淀粉质燃料乙醇发酵胁迫及菌株耐受性改造[J]. 精细化工, 2019, 36(4): 568-574. |
| WANG X Y, QIN L, LIU H, et al. Research progress of starchy fuel ethanol fermentation and the tolerance of Saccharomyces cerevisiae [J]. Fine Chemicals, 2019, 36(4): 568-574. | |
| 60 | PHAN A P H, NGO T T, LENHOFF H M. Spectrophotometric assay for lysine decarylase[J]. Analytical Biochemistry, 1982, 120(1): 193-197. |
| 61 | 夏冰, 丁子元, 郑晓卫, 等. 植物乳杆菌和菌剂及其在生物胺降解、黄酒生产中的应用: CN111254101B[P]. 2020-07-28. |
| XIA B, DING Z Y, ZHENG X W, et al. Lactobacillus plantarum, microbial inoculum and application of Lactobacillus plantarum and microbial inoculum in biogenic amine degradation and yellow rice wine production: CN111254101B[P]. 2020-07-28. | |
| 62 | 王德昌, 明明, 周维广. 分光光度法测定高级醇[J]. 啤酒科技, 2005(3): 37-38. |
| WANG D C, MING M, ZHOU W G. Spectrophotometric determination of higher alcohols[J]. Beer Science and Technology, 2005(3): 37-38. | |
| 63 | 杨鑫, 孙浩轩, 何伟, 等. 一步法制备四甲基吡嗪用反应装置: CN218077901U[P]. 2022-12-20. |
| YANG X, SUN H X, HE W, et al. Reaction device for preparing tetramethylpyrazine by one-step method: CN218077901U[P]. 2022-12-20. | |
| 64 | 丁子元, 杨鑫, 靳喜庆, 等. 枯草芽孢杆菌、菌剂、应用及制备四甲基吡嗪的方法: CN115386525B[P]. 2023-01-31. |
| DING Z Y, YANG X, JIN X Q, et al. Bacillus subtilis, fungicide, application and method for preparing tetramethylpyrazine: CN115386525B[P]. 2023-01-31. | |
| 65 | KONG C L, LI A H, SU J, et al. Flavor modification of dry red wine from Chinese spine grape by mixed fermentation with Pichia fermentans and S. cerevisiae [J]. LWT, 2019, 109: 83-92. |
| 66 | 刘沛通, 丁子元, 于庆泉, 等. 优良本土酿酒酵母的酿造特性研究[J]. 中国食品学报, 2023, 23(1), 204-215. |
| LIU P T, DING Z Y, YU Q Q, et al. Studies on oenological characteristics of high-quality Chinese indigenous Saccharomyces cerevisiae strains[J]. Journal of Chinese Institute of Food Science and Technology, 2023, 23(1), 204-215. | |
| 67 | 刘沛通, 丁子元, 郑晓卫, 等. 酿酒酵母和菌剂以及它们在制备发酵产品特别是怀涿盆地葡萄酒酿造中的应用: CN111961603B[P]. 2021-01-01. |
| LIU P T, DING Z Y, ZHENG X W, et al. Saccharomyces cerevisiae and microbial agent as well as application thereof to preparation of fermented product and particularly brewing of wine in Huazhuo Basin: CN111961603B[P]. 2021-01-01. | |
| 68 | 郑晓卫, 刘沛通, 李泽福, 等. 抗逆性优良的空间育种酿酒酵母及其应用: CN115651852B[P]. 2023-04-11. |
| ZHENG X W, LIU P T, LI Z F, et al. Spatial breeding Saccharomyces cerevisiae with excellent stress resistance and application thereof: CN115651852B[P]. 2023-04-11. | |
| 69 | 王千, 白杰, 江会锋. 合成生物学酶改造设计技术的研究进展[J]. 生命科学, 2021, 33(12): 1493-1501. |
| WANG Q, BAI J, JIANG H F. Research progress on technologies of enzyme engineering and design in synthetic biology[J]. Chinese Bulletin of Life Sciences, 2021, 33(12): 1493-1501. | |
| 70 | 赵聪敏. 甜菊糖苷单体分离、甜味特性及应用研究[D]. 邯郸: 河北工程大学, 2021. |
| ZHAO C M. Isolation of stevia glycoside, sweetness characteristics and application[D]. Handan: Hebei University of Engineering, 2021. | |
| 71 | 祁飞, 刘瑞敏, 张真真. 一种通过易错PCR技术及高通量筛选提高葡萄糖基转移酶EUGT11酶活方法: CN113584016A[P]. 2021-11-02. |
| QI F, LIU R M, ZHANG Z Z. Method for improving enzyme activity of glucosyltransferase EUGT11 through error-prone PCR technology and high-throughput screening: CN113584016A[P]. 2021-11-02. | |
| 72 | 谭剑, 佟毅, 赵国淼, 等. 玉米赤霉烯酮浓度及其降解酶酶活力的测定方法以及玉米赤霉烯酮降解菌的筛选方法: CN112577930A[P]. 2021-03-30. |
| TAN J, TONG Y, ZHAO G M, et al. Zearalenone concentration and zearalenone degrading enzyme activity determination method and zearalenone degrading bacterium screening method: CN112577930A[P]. 2021-03-30. | |
| 73 | 赵国淼, 佟毅, 谭剑, 等. 测定呕吐毒素浓度的方法及其应用: CN114813895A[P]. 2022-07-29. |
| ZHAO G M, TONG Y, TAN J, et al. Method for determining vomitoxin concentration and application thereof: CN114813895A[P]. 2022-07-29. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | Huan LIU, Qiu CUI. Advances and applications of ambient ionization mass spectrometry in screening of microbial strains [J]. Synthetic Biology Journal, 2023, 4(5): 980-999. |
| [3] | zhiqiang ZHANG, Yang ZHANG, Weibao QIU, Hairong ZHENG. Progress and prospect of ultrasonic liquid transfer and low-volume liquid transfer technology [J]. Synthetic Biology Journal, 2023, 4(5): 916-931. |
| [4] | Zhehui HU, Juan XU, Guangkai BIAN. Application of automated high-throughput technology in natural product biosynthesis [J]. Synthetic Biology Journal, 2023, 4(5): 932-946. |
| [5] | MA Cui, YANG Fan, ZHANG Juntai, HE Kai. Multifunction microplate reader for automated foundry platform [J]. Synthetic Biology Journal, 2023, 4(5): 1036-1049. |
| [6] | WU Yujie, LIU Xinxin, LIU Jianhui, Yang Kaiguang, SUI Zhigang, ZHANG Lihua, ZHANG Yukui. Research progress of strain screening and quantitative analysis of key molecules based on high-throughput liquid chromatography and mass spectrometry [J]. Synthetic Biology Journal, 2023, 4(5): 1000-1019. |
| [7] | Weitong QIN, Guangyu YANG. Research and application progress of microdroplets high throughput screening methods [J]. Synthetic Biology Journal, 2023, 4(5): 966-979. |
| [8] | LU Hui, ZHANG Fangli, HUANG Lei. Establishment of iBioFoundry for synthetic biology applications [J]. Synthetic Biology Journal, 2023, 4(5): 877-891. |
| [9] | Yi YANG, Yufeng MAO, Chunhe YANG, Meng WANG, Xiaoping LIAO, Hongwu MA. Recent progress in computational tools for designing editing sequences used in microbial genetic manipulations [J]. Synthetic Biology Journal, 2023, 4(1): 30-46. |
| [10] | Ran TU, Shixin LI, Haoni LI, Meng WANG. Advances and applications of droplet-based microfluidics in evolution and screening of engineered microbial strains [J]. Synthetic Biology Journal, 2023, 4(1): 165-184. |
| [11] | Ting ZHANG, Mengtian LENG, Fan JIN, Hai YUAN. Overview on platform for synthetic biology research at Shenzhen [J]. Synthetic Biology Journal, 2022, 3(1): 184-194. |
| [12] | Yadong CHU, Zongbao ZHAO. Designing and application of small-scale integrated automated liquid handling system [J]. Synthetic Biology Journal, 2022, 3(1): 195-208. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||