|
||
|
Application and prospect of live cell DNA-based molecular recorders in cell lineage tracing
Synthetic Biology Journal
2025, 6 (3):
651-668.
DOI: 10.12211/2096-8280.2024-082
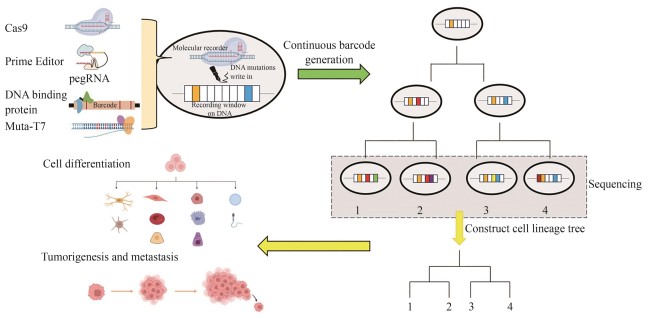
Tracing the division and differentiation history of cells is a critical issue in organismal development and cancer research. Live cell DNA-based molecular recorders, a synthetic system that induces heritable DNA variations, offers an innovative approach for reconstructing cell lineage histories. As a representative for the new generation of cell lineage tracing method, this system can be integrated with high-throughput single-cell sequencing and multi-omics analysis, enabling the reconstruction of developmental differentiation pathways of cells and the phylogenetic trees of tumorigenesis as well. Live cell DNA-based molecular recorders serve as an effective platform for exploring these core biological processes. This review systematically analyzes the technological evolution of Cas9-based molecular recorders in lineage tracing since 2016 and its applications, while also analyzing the research trends of some novel molecular recorders and evaluating their advantages and limitations. Since 2016, molecular recorders based on the CRISPR-Cas9 system have made significant progress and gradually become the mainstream technology in this field. However, Cas9-based molecular recorders still suffer from several inherent limitations, such as the low lineage resolution due to insufficient editing efficiencies, the loss of recorded information caused by DNA double-strand breaks, and potential lineage merging due to barcode homoplasy. These limitations pose challenges for researchers to explore and develop new types of molecular recorders as more efficient and precise tools for cell lineage tracing. Novel molecular recorders based on new principles, such as prime editors, DNA-binding protein-fused base editors, and T7 RNA polymerase-fused base editors, can avoid DNA double-strand breaks and record information through base substitutions rather than deletions. Compared to the Cas9 system, they exhibit unique advantages but also come with potential risks and challenges. Prime editors can record information in a temporal sequential manner, but off-target effects remain a concern. DNA-binding protein-fused base editors offer high editing efficiencies and specificities, but their effectiveness across different cell types requires further exploration. T7 RNA polymerase-fused base editors have achieved success in in vivo directed evolution systems, but their application in mammalian systems is still limited. In the future, the research of DNA-based molecular recorders should focus on optimizing editing efficiency, reducing information loss, improving lineage recovery efficiency, and exploring their application potentials in complicated biological systems.
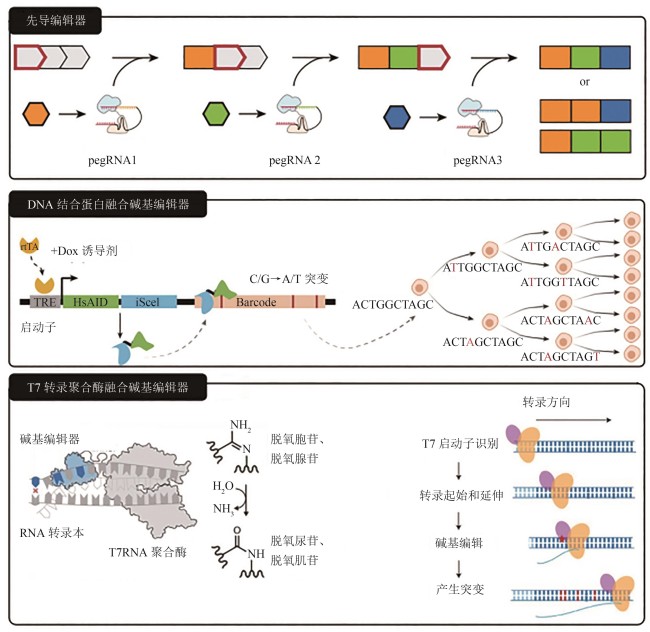
Fig. 3
Working principles for three novel molecular recorders[
(pegRNA—prime editing guide RNA;TRE—a promoter that can be activated by rtTA in the presence of DOX; HsAID—base-editor;iSceI—DNA binding protein; rtTA—inducible system)
Extracts from the Article
进行体内的DNA分子记录的能力并不是CRISPR-Cas系统所独有的。自2020年左右开始,新的基因编辑技术得到广泛关注,目前已经产生了一些不同原理的基于DNA的分子记录器,都具有应用于细胞谱系追踪的前景,并有潜力克服基于Cas9的分子记录器的一些缺点。在本章中,我们将介绍三种有潜力的、基于不同于CRISPR-Cas原理的新型分子记录器(图3),并且将其优势、劣势与基于Cas9的分子记录器进行对比(表2)。
Jay Shendure课题组将先导编辑用于分子记录器的开发和谱系追踪研究。该方法包括一个由部分CRISPR-Cas9靶位点组成的串联阵列,通过短序列插入编辑记录每次编辑的pegRNA身份,同时将引导编辑位置向前移动。这一系统能够保持多达20个连续的序列事件的记录[61](图3)。按序插入式先导编辑细胞历史记录(prime editing cell history recording by ordered insertion,peCHYRON)技术通过逐个插入带有独特条形码的pegRNA,在特定基因组位点插入20 bp的序列,其中包含3 bp的突变作为条形码和17 bp的恒定序列。每次插入都使先前的结合序列失活,从而实现事件的顺序记录[65]。为进一步增强多重记录能力,增强子驱动的基因组多重转录活动系统(enhancer-derived genomic recording of transcriptional activity in multiplex,ENGRAM)方法将多个信号和特异性条形码整合进pegRNA中,从而能够同时捕获多个基因的转录活性[66]。
除了上述两项在发育领域的研究外,同一研究团队也将SMALT体系用于肿瘤的谱系追踪。他们利用SMALT技术追踪小鼠的结直肠癌肿瘤谱系,在单细胞水平上探究了肠道癌变的起源和演化路径[62](图3)。通过分析条形码中的突变模式重建单细胞谱系树,研究人员揭示了大多数肿瘤(66.7%)具有多克隆起源,表明在炎症驱动的肠道肿瘤发生中,多个独立的细胞谱系的并行扩张是常见的[62]。研究人员量化了每个肿瘤的创始祖细胞数量(Np),发现单克隆肿瘤的Np约为1,而多克隆肿瘤的Np在2到33不等,表明早期肿瘤发生可能由多个独立的祖细胞驱动。对于肠道息肉的分析同样支持该结论[68]。研究人员分析了小鼠肠道多个息肉的单细胞谱系,发现所有息肉都表现出多克隆起源,揭示了多克隆起源的复杂性[62]。此外,研究人员比较了单克隆和多克隆肿瘤的突变负担,发现单克隆肿瘤积累了更多的基因组突变,其可能代表了肿瘤进展的晚期阶段[62]。最后,通过谱系发生树估计祖细胞的时间,研究人员推断了息肉起始的克隆扩张时间,并结合小鼠肠上皮细胞的增殖速率,以此推测人类家族性腺瘤性息肉病(FAP)患者的肿瘤起始可能早在婴儿期就已发生[62]。这些重要发现为结直肠癌的早期筛查和干预提供了新视角,指出在多克隆病变阶段介入的潜在机会,可能改善临床结果[62]。
Other Images/Table from this Article
|