|
||
|
Exploration of gene functions and library construction for engineering strains from a synthetic biology perspective
Synthetic Biology Journal
2025, 6 (3):
685-700.
DOI: 10.12211/2096-8280.2024-079
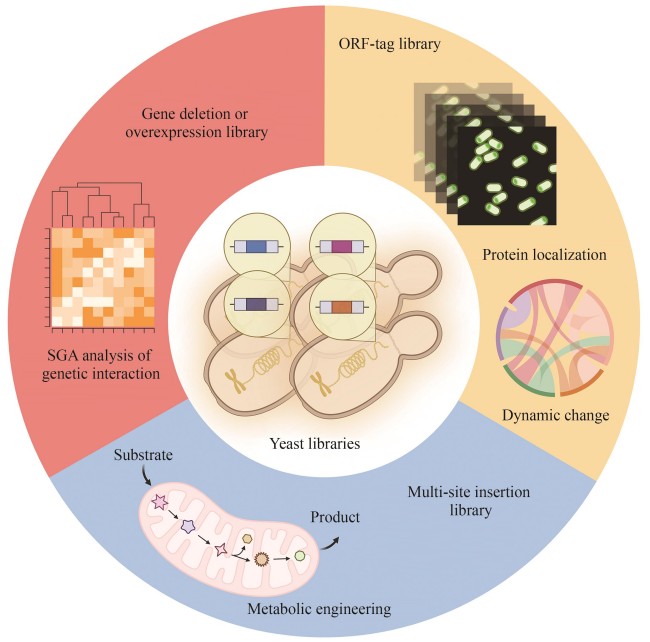
Synthetic biology, as a discipline that designs, constructs, and modifies biological systems to achieve specific functions, is widely applied in biomanufacturing, the biodegradation of environmental pollutants, and drug synthesis. Systematic exploration of gene functions and construction of libraries for engineered strains are driving forces of the development of synthetic biology. These libraries serve as foundational tools for understanding complex biological processes and engineering microorganisms for potential applications. This review focuses on the construction methods and application prospects of various yeast libraries in synthetic biology. With the rapid advancement of genome sequencing and high-throughput screening technologies, microbial libraries, such as those of Saccharomycescerevisiae and Schizosaccharomycespombe, play a pivotal role in systematic research. Yeast libraries, including gene knockout libraries, overexpression libraries, and transposon insertion libraries, provide valuable tools for optimizing gene combinations and designing metabolic pathways, thus promoting applications in metabolic engineering and synthetic biology. These libraries facilitate the development of robust industrial strains, driving improvements in biofuel production, chemical synthesis, and other biotechnological processes. In the environmental field, the screening of modified genes generates strains with pollutant degradation capabilities, contributing to ecological restoration. In drug synthesis, these libraries aid in constructing strains for the efficient production of pharmaceutical compounds, advancing the development of biopharmaceuticals. Despite these successes, there remain challenges in library construction and application, such as the high cost of library generation, difficulty in precise genome editing, and limitation in screening efficiency. In the future, advances in automation, digitization, and novel screening technologies are expected to overcome these barriers, facilitating the rapid construction and efficient screening of yeast libraries. No doubt, synthetic biology holds immense promise, with improvements in library construction and screening processes expected to accelerate the development of sustainable solutions in industrial production, environmental protection, and healthcare, thereby driving innovations in biotechnology.
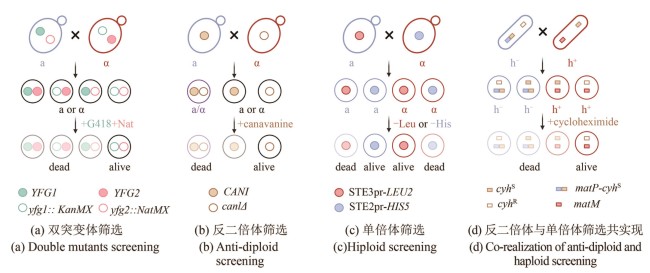
Fig. 2
Schematics for SGA and PEM methods (created by biorender)
[(a) Double mutant selection strategy. Two mating-type cells each carry different resistance markers at their respective mutation sites: KanMX (green hollow circle) and NatMX (pink hollow circle). These allow for the selection of double mutant progeny (outlined in black). YFG (your favorite gene): target gene of interest.(b) Counter-diploid selection strategy. Haploid cells carrying a wild-type CAN1 gene (blue, a-type) and diploid cells (purple) can uptake the toxic analog canavanine and are killed, while can1Δ mutants cannot transport canavanine and thus survive (outlined in black).(c) Haploid selection strategy. A mating-type-specific promoter is used to drive the expression of a nutritional selection marker in the opposite mating type, allowing selection of haploid progeny of a specific mating type. For example, when an a-type parent cell (blue), harboring a gene cassette expressing STE3pr-LEU2 (red solid circle) — which is only active in α-type cell — mates with another cell, only the α-type progeny that inherit this construct (outlined in red with a red solid circle) will survive.(d) Combined counter-diploid and haploid selection strategy. A dominant-lethal resistance gene, cyhS (brown solid square), is inserted near the mat1 mating-type locus to link its expression with a specific haploid phenotype. For example, insertion near matP (blue square) in h- cells (blue) leads to the death of cyhS -containing h- haploids and diploids, enabling selection of h+ haploids.]
Extracts from the Article
2001年,Tong等[23]开发了合成遗传阵列(synthetic genetic array,SGA)方法用于系统构建双基因修饰(敲除或过表达)文库,该方法利用自发的交配途径代替了冗长的转化过程。以构建酿酒酵母单倍双突变体为例(图2),在缺氮条件下诱导两种相反交配类型(MATa和MATα)的细胞交配,这些母本菌株包括以下3个特征:①它们分别在各自的突变位点携带不同抗性标签(KanMX和NatMX)用于筛选双突变体子代[图2(a)]。②在任一母本菌株中携带负筛选型突变(如can1Δ、lyp1Δ等),用于在选择性培养基上反筛选二倍体[图2(b)]。含有野生型CAN1基因的单倍体细胞或二倍体细胞会摄入有毒的刀豆氨酸(canavanine),从而被杀死,而can1Δ突变体无法将刀豆氨酸运转入体内,因此能够存活。③在某一细胞型的母本菌株中,构建另一细胞型特异性启动子与营养缺陷筛选标签的表达盒,用于选择任一性别的单倍体子代细胞[图2(c)]。如在MATa细胞中构建MATα特异型启动子STE2pr驱动his5基因的表达盒,只有当细胞发生交配产孢后,STE2pr-his5才能在MATα子代细胞中表达从而允许其在组氨酸缺陷型培养基上生长;同理,在MATα细胞中预先组装STE3pr-lue2表达盒,由于STE3pr只能在MATa细胞中表达,可以通过亮氨酸缺陷型培养基筛选MATa子代细胞。除了使用细胞型特异性启动子以外,Krogan课题组[24]还在裂殖酵母中开发了一种特定单倍体的偶联筛选策略PEM(pombe epistasis mapper),他们通过将一个显性致死的抗性基因(cyhS)“镶嵌”在交配位点mat1附近,使得某一单倍体表型与抗性基因的表达偶联,在添加环己胺(cycloheximide)的培养基上实现对这一单倍体的反筛[图2(d)]。根据双基因缺失菌株相对于任一单基因缺失菌株的生长情况,可以判断两个基因之间发生的负向或正向相互作用,揭示基因间的冗余或旁路抑制。
Other Images/Table from this Article
|