|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Synthetic genetic circuit engineering: principles, advances and prospects
Synthetic Biology Journal
2025, 6 (1):
45-64.
DOI: 10.12211/2096-8280.2023-096
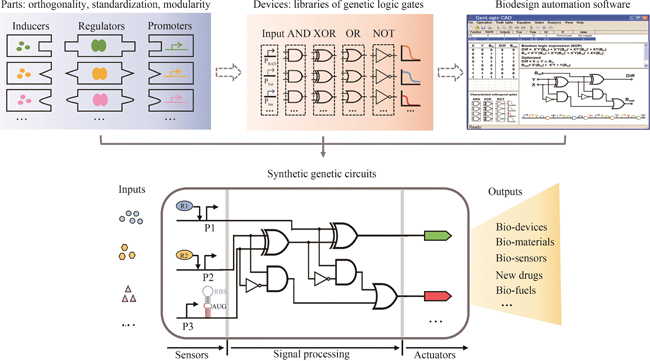
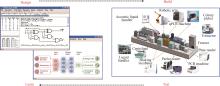
Synthetic genetic circuits are engineered gene networks comprised of redesigned genetic parts for interacting to perform customized functions in cells. With the rapid development of synthetic biology, synthetic genetic circuits have shown significant application potentials in many fields such as biomanufacturing, healthcare and environmental monitoring. However, the efforts to scale up genetic circuits are hindered by the limited number of orthogonal parts, the difficulty of functionally composing large-scale circuits, and the poor predictability of circuit behaviors. A longstanding goal of synthetic biology research is to engineer complex synthetic biological circuits, using modular genetic parts, as we do with electronic circuits. Synthetic biologists have developed various genetic toolboxes and functional assembly methods over the past few decades. Here we present an overview of the latest advances, challenges, and future prospects in genetic circuit engineering from four aspects corresponding to the four key engineering principles for circuit design, i.e. orthogonality, standardization, modularity, and automation. Firstly, the design and construction of orthogonal genetic part libraries are discussed in both prokaryotes and eukaryotes at the levels of DNA replication, transcription, and translation, respectively. Standardized characterization methods and the design of modular genetic parts are subsequently summarized. Furthermore, progress in developing modular genetic circuits are presented, providing new concepts and ways for engineering increasingly large and complex circuits. Finally, how to achieve automated design and building of genetic circuits are addressed from the advances in software, hardware and artificial intelligence, respectively, with an aim to replacing the presently time-consuming manual trial-and-error mode with the iterative "design-build-test-learn" cycle for improved efficiency and predictability of circuit design. The integration of these fundamental principles and the latest advances in information technology such as artificial intelligence and lab automation will accelerate the paradigm shift in genetic circuit engineering and synthetic biology research, making it feasible for designing synthetic lives to meet various customized needs.
Table 1
Design and characterization of the libraries of orthogonal genetic parts
Extracts from the Article
生物正交反应是指能够在生物体系中进行,且不会与天然生物化学过程相互干扰的一类化学反应[13]。在基因线路设计过程中,为了精准地调控细胞内各组分的活性,应尽量减少元件之间的相互干扰,这就是基因线路中的“正交化”。随着合成生物学的快速发展,科学家们在遗传信息表达及生化代谢的不同过程中成功创建了正交元件(图1和表1)。
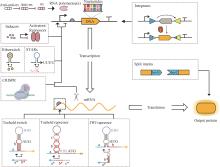
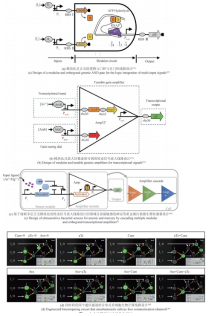
同时,单个细胞的负载有限,组合多种功能细胞,可以提高群体对复杂环境的适应性和鲁棒性,以完成更加复杂的工作。可通过多细胞分布式线路设计,让单个细胞里的线路模块化并进行多个细胞间的级联通信来提高线路的模块性和实现线路的规模化组装设计。相比于基于单种细胞的计算线路,分布式计算不仅可以降低每个细胞中合成线路的体积,减少代谢负担,而且可以利用细胞高度模块化的特性,在不同的细胞群体内重复使用相同的基因元件,降低对正交基因调控元件的依赖。为了演示更复杂的多细胞生物计算功能,研究人员设计了复杂的三输入XOR-AND逻辑门电路。XOR-AND逻辑门电路分别部署在七个不同的大肠杆菌菌株中,由四个通信通道协调。每个菌株包含一个NOR门(cell-1至cell-6)或一个Buffer门(cell-7),这是已知第一个同时利用四个通信通道的生物计算线路[图3(d)][38]。虽然多细胞分布式计算在细菌、酵母和哺乳动物细胞中都已经实现,但它们的规模却并未得到显著的提高。在细菌中最具有代表性的分布式计算线路仍然是基于多细胞或非门的16个2输入逻辑门以及更复杂一些的与门-异或门复合逻辑[3, 38]。而在酵母与哺乳动物细胞中,分布式计算线路的规模也与单细胞计算线路的规模相差不大,最具代表性的是1位全加器程序[90-91]。
Other Images/Table from this Article
|