|
||
|
Synthetic genetic circuit engineering: principles, advances and prospects
Synthetic Biology Journal
2025, 6 (1):
45-64.
DOI: 10.12211/2096-8280.2023-096
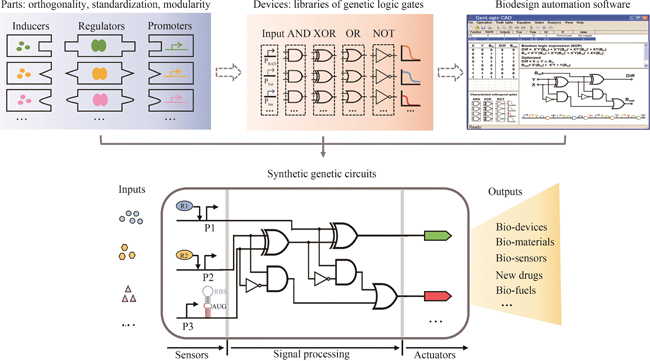
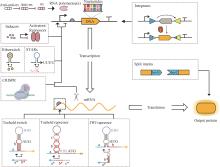
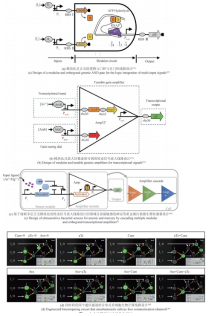
Synthetic genetic circuits are engineered gene networks comprised of redesigned genetic parts for interacting to perform customized functions in cells. With the rapid development of synthetic biology, synthetic genetic circuits have shown significant application potentials in many fields such as biomanufacturing, healthcare and environmental monitoring. However, the efforts to scale up genetic circuits are hindered by the limited number of orthogonal parts, the difficulty of functionally composing large-scale circuits, and the poor predictability of circuit behaviors. A longstanding goal of synthetic biology research is to engineer complex synthetic biological circuits, using modular genetic parts, as we do with electronic circuits. Synthetic biologists have developed various genetic toolboxes and functional assembly methods over the past few decades. Here we present an overview of the latest advances, challenges, and future prospects in genetic circuit engineering from four aspects corresponding to the four key engineering principles for circuit design, i.e. orthogonality, standardization, modularity, and automation. Firstly, the design and construction of orthogonal genetic part libraries are discussed in both prokaryotes and eukaryotes at the levels of DNA replication, transcription, and translation, respectively. Standardized characterization methods and the design of modular genetic parts are subsequently summarized. Furthermore, progress in developing modular genetic circuits are presented, providing new concepts and ways for engineering increasingly large and complex circuits. Finally, how to achieve automated design and building of genetic circuits are addressed from the advances in software, hardware and artificial intelligence, respectively, with an aim to replacing the presently time-consuming manual trial-and-error mode with the iterative "design-build-test-learn" cycle for improved efficiency and predictability of circuit design. The integration of these fundamental principles and the latest advances in information technology such as artificial intelligence and lab automation will accelerate the paradigm shift in genetic circuit engineering and synthetic biology research, making it feasible for designing synthetic lives to meet various customized needs.

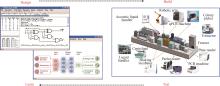
Fig. 2
Design portfolio and the web architecture of the BioPartsDB platform[
(a) A simplified diagram showing the information flow of the database platform. Arrows indicate the intended user browsing along the platform's webpages. Increasingly stacked panels indicate the higher number of pages in each section and consequently the more detailed level of information. (b) A web page with in-depth description of the information, performance, and characterization conditions for a specific genetic part. (c) A table for parts with a brief description and data of their key performance.
Extracts from the Article
同时,在基因元件的表征方面,目前缺少标准化的评定方法,这就造成已有的如Registry for Standard Biological Parts和SynBioHub[72]基因元件存储库中的各元件之间无法进行准确的定量比较。研究者只能在同时表征大量基因元件的研究文献中获得具有可比性的数据,但归纳总结这类研究,并获得理想的数据十分低效耗时。为此,本团队对已经过实验验证和文献报道的标准基因元器件及其表征数据进行了系统的分类,建立了BioPartsDB(
Other Images/Table from this Article
|