合成生物学 ›› 2022, Vol. 3 ›› Issue (1): 238-251.DOI: 10.12211/2096-8280.2021-082
• 研究论文 • 上一篇
ExoCET-BAC策略高效抓取和组装高AT含量基因组大片段
姜婵娟1,2, 崔天琦1, 孙洪娈1, 焦念志2, 符军1, 张友明1, 王海龙1
- 1.山东大学微生物技术研究院,微生物技术国家重点实验室,山东 青岛 266237
2.山东大学海洋研究院,山东 青岛 266237
-
收稿日期:2021-08-06修回日期:2021-08-28出版日期:2022-02-28发布日期:2022-03-14 -
通讯作者:张友明,王海龙 -
作者简介:姜婵娟 (1990—),女,博士研究生。研究方向为微生物基因编辑与药物合成生物学等。E-mail:chanjuanjiang@163.com张友明 (1964—),男,教授,博士生导师。研究方向为研究方向为基因组编辑与合成生物学等。E-mail:zhangyouming@sdu.edu.cn王海龙 (1984—),男,教授,博士生导师。研究方向为微生物基因编辑与药物合成生物学等。E-mail:wanghailong@sdu.edu.cn -
基金资助:山东省泰山学者项目(tsqn201812008);山东大学齐鲁青年学者项目
Efficient capture and assembly of AT-rich genomic fragments using ExoCET-BAC strategy
JIANG Chanjuan1,2, CUI Tianqi1, SUN Hongluan1, JIAO Nianzhi2, FU Jun1, ZHANG Youming1, WANG Hailong1
- 1.State Key Laboratory of Microbial Technology,Institute of Microbial Technology,Shandong University,Qingdao 266237,Shandong,China
2.Institute of Marine Science and Technology,Shandong University,Qingdao 266237,Shandong,China
-
Received:2021-08-06Revised:2021-08-28Online:2022-02-28Published:2022-03-14 -
Contact:ZHANG Youming, WANG Hailong
摘要:
基因克隆是解析基因功能的重要手段,但仍有很多基因难以克隆,比如高AT含量(>60%)基因组来源的DNA。ExoCET克隆技术通过联合核酸外切酶介导的体外同源重组和大肠杆菌RecET重组酶介导的细胞内同源重组,不仅能从微生物基因组中靶向抓取>100 kb的大片段,而且能高效组装>13个DNA片段,是基因克隆的有力工具,迄今未有利用ExoCET技术从AT含量>63%的基因组克隆大片段的报道。本研究以AT含量为69%的海洋单细胞光合蓝细菌原绿球藻MIT 9301菌株的基因组为研究对象,探究了利用ExoCET技术进行高AT含量基因组大片段克隆的最佳条件。结果显示:①在核酸外切酶介导的体外同源重组时使用Gibson体系较T4聚合酶体系能获得更高的克隆效率;②载体应选择单拷贝的细菌人工染色体(BAC),多拷贝质粒载体会导致克隆失败;③ExoCET可以从原绿球藻基因组上抓取>80 kb的大片段,并且能以100%的正确率组装11个3 kb的DNA片段;④可以一步同时抓取4个7~20 kb的基因组大片段。大规模基因组测序显示高AT含量生物占比超过30%,该研究建立的ExoCET-BAC策略将为高AT含量生物的基因组功能研究提供高效使能技术。
中图分类号:
引用本文
姜婵娟, 崔天琦, 孙洪娈, 焦念志, 符军, 张友明, 王海龙. ExoCET-BAC策略高效抓取和组装高AT含量基因组大片段[J]. 合成生物学, 2022, 3(1): 238-251.
JIANG Chanjuan, CUI Tianqi, SUN Hongluan, JIAO Nianzhi, FU Jun, ZHANG Youming, WANG Hailong. Efficient capture and assembly of AT-rich genomic fragments using ExoCET-BAC strategy[J]. Synthetic Biology Journal, 2022, 3(1): 238-251.
| Characteristics | Source | |
|---|---|---|
| Strains | ||
| E. coli GB2005 | E. coli DH10B derivates in which fhuA::IS2, ΔrecET, ΔybcC; endogenous recET and DLP12 phage ybcC genes are deleted, and fhuA is mutated to make it resistant to T1 phage | Laboratory stock |
| E. coli GB05-dir | GB05 derivates expressing full-length RecE/RecT under the arabinose-inducible PBAD promoter | Laboratory stock |
| Plasmids | ||
| pSC101-BAD-ETgA-tet | Full-length RecE/RecT inducible (PBAD promoter) expression plasmid contains a temperature-sensitive pSC101 origin and a tetracycline resistance gene. | Laboratory stock |
| pBR322-amp-ccdB-rpsL | A pBR322 plasmid with the ampicillin resistance gene, kanamycin resistance gene, and counterselection genes ccdB and rpsl. | Laboratory stock |
| pBeloBAC11-hygccdB | A bacterial artificial chromosome (BAC) with the chloramphenicol resistance gene, hygromycin resistance gene and ccdB gene | Laboratory stock |
表1 本研究所用的菌株和质粒
Tab. 1 Strains and plasmids used in this study
| Characteristics | Source | |
|---|---|---|
| Strains | ||
| E. coli GB2005 | E. coli DH10B derivates in which fhuA::IS2, ΔrecET, ΔybcC; endogenous recET and DLP12 phage ybcC genes are deleted, and fhuA is mutated to make it resistant to T1 phage | Laboratory stock |
| E. coli GB05-dir | GB05 derivates expressing full-length RecE/RecT under the arabinose-inducible PBAD promoter | Laboratory stock |
| Plasmids | ||
| pSC101-BAD-ETgA-tet | Full-length RecE/RecT inducible (PBAD promoter) expression plasmid contains a temperature-sensitive pSC101 origin and a tetracycline resistance gene. | Laboratory stock |
| pBR322-amp-ccdB-rpsL | A pBR322 plasmid with the ampicillin resistance gene, kanamycin resistance gene, and counterselection genes ccdB and rpsl. | Laboratory stock |
| pBeloBAC11-hygccdB | A bacterial artificial chromosome (BAC) with the chloramphenicol resistance gene, hygromycin resistance gene and ccdB gene | Laboratory stock |
| Oligos | Sequence (5′→3′) | Purpose | Template |
|---|---|---|---|
| pBR322-5pieces-1 | TTTTAGTTTATTTTTAAGACTTTTTAATACTGGATCATTCGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 5-piece DNA assembly | pBR322-amp-ccdB-rpsL |
| pBR322-2 | GGGAACTGTGGAATTCTTAAATTAAATACCTTGTCGAGGTGTTTAAACGGTGTGGTAGCTCGCGTATT | ||
| F1-1 | ACCTCGACAAGGTATTTAAT | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F1-2 | CAACATTCTTTCTTGCGATT | ||
| F2-1 | CATACCAATATTTATTATCT | ||
| F2-2 | AGACCTAAATAAATTAAATT | ||
| F3-1 | ATTTTGATTTATTTGGTTTG | ||
| F3-2 | TAGCAAACAATACAAAAACG | ||
| F4-1 | CTTTATTTAGAGAATTTAAT | ||
| F4-2 | TCTGTAATTCTTGCTTTTGCTCTACC | ||
| pBR322-7pieces-1 | TTAAAAAAACAACAGGTAGAGCAAAAGCAAGAATTACAGAGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 7-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F5-1 | TGTTGCTTGTGATGAGGCTG | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F5-2 | AGATTTAAGAAGGATTTTTG | ||
| F6-1 | GAAATTTTAATTGCCGATTT | ||
| F6-2 | CACATCCTGCACAATGAAAA | ||
| pBR322-9pieces-1 | AATGAGAAGAGAAAAGGGAATTTTCATTGTGCAG GATGTGGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 9-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F7-1 | ATAATGAGAAGAGAAAAGGGA | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F7-2 | ATTTTGAAGGCCTGGAATTA | ||
| F8-1 | GCAAAACTACTTTGCTTAAT | ||
| F8-2 | TTTTGATTTAAGTAAAAGAT | ||
| pBR322-11pieces-1 | TAATTCTTTATCCAATGTTTATCTTTTACTTAAATCA AAAGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 11-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F9-1 | AGTAATTCTTTATCCAATGT | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F9-2 | TCAATCTTTACTAAAAAAAT | ||
| F10-1 | ATTCTTGAATAATTTTACTCT | ||
| F10-2 | GAATGATCCAGTATTAAAAAG | ||
| BAC-11pieces-1 | TTTTAGTTTATTTTTAAGACTTTTTAATACTGGATCATTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector for 11-piece DNA assembly. | pBeloBAC11-hygccdB |
| BAC-11pieces-2 | GGGAACTGTGGAATTCTTAAATTAAATACCTTGTCGAGGTGTTTAAACCGGGTACCGAGCTCGAATTCG | ||
| BAC-49kb-1 | CCTTAAAAGATTGATTATTTTTCAACCATTATTCAATTTTCCCATCATAGATGGCATTAGTTATTCTCCCAGGTTTAAACGCGGCCGCCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 49 kb fragment | |
| BAC-49kb-2 | GAATTCGAGCTCGGTACCCGGCGGCCGCGGTTTAAACATAGATTGTGTATTGGCGTGTTTAAAAAAAAAATGGAAGAATGAATTAATACAGATTTAGTAACTAATCTA | ||
| BAC-21kb-1 | TCAATGTCCAGAACTTAAAGAGGTATTACGCCTCATTGAAATAGGTCACTTTAGCAATGGGGATAAAGAATTGTTTAAACGCGGCCGCCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 21 kb fragment | |
| BAC-21kb-2 | GAATTCGAGCTCGGTACCCGGCGGCCGCGTTTAAACATTGCTAATCCCCTCCTTGCATCTTGCAAAGATACTTGTTGAGGAGTAGTGACAACTATAGCTCCAGAAATA | ||
| BAC-82kb-1 | TTTTACCTGGCATATCTGGCGGAATAAATACTGACATATTTTCTACAATTCCTAGTAAAGGTACTCCGAGTTGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 82 kb fragment | |
| BAC-82kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACAATCTTATTGAAGACATTAGTGAGGAAAAAAAATTACCTCCTAATATCGTTGAAGCAGCCTTGCGCGAAGCT | ||
| BAC-65kb-1 | ATTTACCAAATCCAAGGAATGAAGCAGTAGAAAATGATCTAATTGTTGATAATAAATGCTTTATAGAATTAGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 65 kb fragment | |
| BAC-65kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTTTATTAAATAGCTTGACCGGACATGATCCATTCTTTGTTATGGCGGACTTTGAAGACTACCTAAACAAAC | ||
| BAC-22kb-1 | TATGGGATCAAAAAGAAATCCCAAGAATTGCTCAATTGGTATATTTGGAGTTAATTACGACGGGACATGTTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 22 kb fragment | |
| BAC-22kb -2 | GAATTCGAGCTCGGTACCCGGTTTAAACCCAACTACTCTTGATAATGGCCTATTAGAAGAAGTTGTTGAAGTTGCTAAAAAATACTCCAATAGATGTGAT | ||
| BAC-18kb-A1 | ATCAAATAAGGTATCTTTACTCAATTTGTAAAAAAGTTACTTACTATCCGATAATAGGATTAATCTGAGGCTGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 18 kb-A fragment | |
| BAC-18kb-A2 | GAATTCGAGCTCGGTACCCGGTTTAAACAATGATGAATGCCATAAGATTTATAAATATAAAATGTGCCAGGTTTGCCAAATAATGATTTGTTTGATTCAG | ||
| BAC-54kb-1 | CAAGGTAACAACTGCGAGAGTTAGAATAAGAACGTAAGCAAAAGCTTCCATAAGATTAAGTAATTAAGTTTAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 54 kb fragment | |
| BAC-54kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACACACATTTACACTTAGTGTTTGAATTTCAAGTCTATTTCGGGTTGAATTAAATTGTTTTTTTATAGTAGTTA | ||
| BAC-25kb-1 | CACTGTAATTAGTGAAATAAACCCCTTCTGTCTCCTTTTTGATTTCTTCAGTCCATCTTTCTTGTGGAGCAAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 25 kb fragment | |
| BAC-25kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACGGTCAACGAATTTCTAACGGTTCCGATTTCGACGAACAAACCGGTAAATTAAAAGAAGGGAACAAATCTTTA | ||
| BAC-20kb-1 | TCCCATAACATCAATTAAAAGCGCTATTTCTTATAAGAAATCTATTATTGAAGCGTTGCCAGAAAGTTCTAAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 20 kb fragment | |
| BAC-20kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACAGAGGAGAACGAAAAAAAGGTAGTTCTCTTGTCACAGGATCTGAGGTGCAATCTCAGGCCAGTGGTGCAAGC | ||
| BAC-18kb-B1 | AAAATTTATATTAGAAACCATAGTCTCTTTAGTTTTACTTTTATTAGTTATTTTTAAATCAATAATTAATTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 18kb-B fragment | |
| BAC-18kb-B2 | GAATTCGAGCTCGGTACCCGGTTTAAACAGGGTTCTATAAAAGTTTTAATTAAATCAATAAGTGTTATTTGAAAATCCACCAAAAAATATAGAGAGCTTA | ||
| BAC-10kb-1 | TTGAGATGTTAGATATTGTAGTTAACAAAAAAAGAGAAGTGTTTAACGGTTTAAATAAACATAGGGACTATAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 10 kb fragment | |
| BAC-10kb-2 | TAAATATTAATGATATTTCCTTTTATTCAATCCTTTACTATTTGAGCGGTGCAATATGCCCATCTTTGGTGTGTTTAAACCGGGTACCGAGCTCGAATTC | ||
| BAC-11kb-1 | ACGGAATGCAAGAGGATGGTGCTAGTAACGAAACATATACAGCATTAAACGGTTTCTATTCATTTGATAACGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 11 kb fragment | |
| BAC-11kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCGTTATCAAATGAATAATAACCATTAAATGCCATGTATGTATCGTTGTCGTTACCATCTTCGATGTTTGCGT | ||
| BAC-7kb-1 | TACAGGCATTCCGGCAGTTTTAAAATTACTGCCGGAAATGTTTCCTCCTGATTTCTCAAGAGCTGGAACTTGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 7 kb fragment | |
| BAC-7kb-2 | GAAGTACGCTAATGTTTCGTCCACATTTGCGTCAGCAACATCGAGATCTCCGACTTCATAACCAGCACTGATGTTTAAACCGGGTACCGAGCTCGAATTC | ||
| BAC-12kb-1 | TAAAAGAATTGCAAAATGAGTCATATATAATTAAGAAAAATGATCAGGAAGGGGTTATAAAAAGTAATACTGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 12 kb fragment | |
| BAC-12kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTTTAATAGACTCTTTGGCAATGAGTATCGCAGGAACTTCGATTTCTGTTTTTTTATCTTTACTTCTATGTT | ||
| BAC-67kb-1 | TCCTTGATTTAATGTCCAGTTGGGATAGTTATTTACCTAAAATGAAAAAATATAAAAAAGTTATAGGACATGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 67 kb fragment | |
| BAC-67kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACGGATATCACTCTAAAGATATAAGGAATTTTGAAAATATCCCAAAAATCAATAAGTTGATGTTTGAGATAGCT | ||
| BAC-50kb-1 | GAGTTCAGACCCAGCAGATATTCCTCGATATAAAGCAGCCTAAATTACAAAAAGCTTGAAAACTATATAAATGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 50 kb fragment | |
| BAC-50kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTACCCAGGGAGAAGACATATCATGTAAATTAGTCTGAAATCCCATTTCTTCTAATTCATAGCCTATTGGAT |
表2 本研究所用寡核苷酸序列
Tab. 2 Oligonucleotide sequences used in this study
| Oligos | Sequence (5′→3′) | Purpose | Template |
|---|---|---|---|
| pBR322-5pieces-1 | TTTTAGTTTATTTTTAAGACTTTTTAATACTGGATCATTCGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 5-piece DNA assembly | pBR322-amp-ccdB-rpsL |
| pBR322-2 | GGGAACTGTGGAATTCTTAAATTAAATACCTTGTCGAGGTGTTTAAACGGTGTGGTAGCTCGCGTATT | ||
| F1-1 | ACCTCGACAAGGTATTTAAT | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F1-2 | CAACATTCTTTCTTGCGATT | ||
| F2-1 | CATACCAATATTTATTATCT | ||
| F2-2 | AGACCTAAATAAATTAAATT | ||
| F3-1 | ATTTTGATTTATTTGGTTTG | ||
| F3-2 | TAGCAAACAATACAAAAACG | ||
| F4-1 | CTTTATTTAGAGAATTTAAT | ||
| F4-2 | TCTGTAATTCTTGCTTTTGCTCTACC | ||
| pBR322-7pieces-1 | TTAAAAAAACAACAGGTAGAGCAAAAGCAAGAATTACAGAGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 7-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F5-1 | TGTTGCTTGTGATGAGGCTG | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F5-2 | AGATTTAAGAAGGATTTTTG | ||
| F6-1 | GAAATTTTAATTGCCGATTT | ||
| F6-2 | CACATCCTGCACAATGAAAA | ||
| pBR322-9pieces-1 | AATGAGAAGAGAAAAGGGAATTTTCATTGTGCAG GATGTGGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 9-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F7-1 | ATAATGAGAAGAGAAAAGGGA | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F7-2 | ATTTTGAAGGCCTGGAATTA | ||
| F8-1 | GCAAAACTACTTTGCTTAAT | ||
| F8-2 | TTTTGATTTAAGTAAAAGAT | ||
| pBR322-11pieces-1 | TAATTCTTTATCCAATGTTTATCTTTTACTTAAATCA AAAGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 11-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F9-1 | AGTAATTCTTTATCCAATGT | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F9-2 | TCAATCTTTACTAAAAAAAT | ||
| F10-1 | ATTCTTGAATAATTTTACTCT | ||
| F10-2 | GAATGATCCAGTATTAAAAAG | ||
| BAC-11pieces-1 | TTTTAGTTTATTTTTAAGACTTTTTAATACTGGATCATTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector for 11-piece DNA assembly. | pBeloBAC11-hygccdB |
| BAC-11pieces-2 | GGGAACTGTGGAATTCTTAAATTAAATACCTTGTCGAGGTGTTTAAACCGGGTACCGAGCTCGAATTCG | ||
| BAC-49kb-1 | CCTTAAAAGATTGATTATTTTTCAACCATTATTCAATTTTCCCATCATAGATGGCATTAGTTATTCTCCCAGGTTTAAACGCGGCCGCCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 49 kb fragment | |
| BAC-49kb-2 | GAATTCGAGCTCGGTACCCGGCGGCCGCGGTTTAAACATAGATTGTGTATTGGCGTGTTTAAAAAAAAAATGGAAGAATGAATTAATACAGATTTAGTAACTAATCTA | ||
| BAC-21kb-1 | TCAATGTCCAGAACTTAAAGAGGTATTACGCCTCATTGAAATAGGTCACTTTAGCAATGGGGATAAAGAATTGTTTAAACGCGGCCGCCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 21 kb fragment | |
| BAC-21kb-2 | GAATTCGAGCTCGGTACCCGGCGGCCGCGTTTAAACATTGCTAATCCCCTCCTTGCATCTTGCAAAGATACTTGTTGAGGAGTAGTGACAACTATAGCTCCAGAAATA | ||
| BAC-82kb-1 | TTTTACCTGGCATATCTGGCGGAATAAATACTGACATATTTTCTACAATTCCTAGTAAAGGTACTCCGAGTTGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 82 kb fragment | |
| BAC-82kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACAATCTTATTGAAGACATTAGTGAGGAAAAAAAATTACCTCCTAATATCGTTGAAGCAGCCTTGCGCGAAGCT | ||
| BAC-65kb-1 | ATTTACCAAATCCAAGGAATGAAGCAGTAGAAAATGATCTAATTGTTGATAATAAATGCTTTATAGAATTAGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 65 kb fragment | |
| BAC-65kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTTTATTAAATAGCTTGACCGGACATGATCCATTCTTTGTTATGGCGGACTTTGAAGACTACCTAAACAAAC | ||
| BAC-22kb-1 | TATGGGATCAAAAAGAAATCCCAAGAATTGCTCAATTGGTATATTTGGAGTTAATTACGACGGGACATGTTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 22 kb fragment | |
| BAC-22kb -2 | GAATTCGAGCTCGGTACCCGGTTTAAACCCAACTACTCTTGATAATGGCCTATTAGAAGAAGTTGTTGAAGTTGCTAAAAAATACTCCAATAGATGTGAT | ||
| BAC-18kb-A1 | ATCAAATAAGGTATCTTTACTCAATTTGTAAAAAAGTTACTTACTATCCGATAATAGGATTAATCTGAGGCTGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 18 kb-A fragment | |
| BAC-18kb-A2 | GAATTCGAGCTCGGTACCCGGTTTAAACAATGATGAATGCCATAAGATTTATAAATATAAAATGTGCCAGGTTTGCCAAATAATGATTTGTTTGATTCAG | ||
| BAC-54kb-1 | CAAGGTAACAACTGCGAGAGTTAGAATAAGAACGTAAGCAAAAGCTTCCATAAGATTAAGTAATTAAGTTTAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 54 kb fragment | |
| BAC-54kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACACACATTTACACTTAGTGTTTGAATTTCAAGTCTATTTCGGGTTGAATTAAATTGTTTTTTTATAGTAGTTA | ||
| BAC-25kb-1 | CACTGTAATTAGTGAAATAAACCCCTTCTGTCTCCTTTTTGATTTCTTCAGTCCATCTTTCTTGTGGAGCAAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 25 kb fragment | |
| BAC-25kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACGGTCAACGAATTTCTAACGGTTCCGATTTCGACGAACAAACCGGTAAATTAAAAGAAGGGAACAAATCTTTA | ||
| BAC-20kb-1 | TCCCATAACATCAATTAAAAGCGCTATTTCTTATAAGAAATCTATTATTGAAGCGTTGCCAGAAAGTTCTAAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 20 kb fragment | |
| BAC-20kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACAGAGGAGAACGAAAAAAAGGTAGTTCTCTTGTCACAGGATCTGAGGTGCAATCTCAGGCCAGTGGTGCAAGC | ||
| BAC-18kb-B1 | AAAATTTATATTAGAAACCATAGTCTCTTTAGTTTTACTTTTATTAGTTATTTTTAAATCAATAATTAATTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 18kb-B fragment | |
| BAC-18kb-B2 | GAATTCGAGCTCGGTACCCGGTTTAAACAGGGTTCTATAAAAGTTTTAATTAAATCAATAAGTGTTATTTGAAAATCCACCAAAAAATATAGAGAGCTTA | ||
| BAC-10kb-1 | TTGAGATGTTAGATATTGTAGTTAACAAAAAAAGAGAAGTGTTTAACGGTTTAAATAAACATAGGGACTATAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 10 kb fragment | |
| BAC-10kb-2 | TAAATATTAATGATATTTCCTTTTATTCAATCCTTTACTATTTGAGCGGTGCAATATGCCCATCTTTGGTGTGTTTAAACCGGGTACCGAGCTCGAATTC | ||
| BAC-11kb-1 | ACGGAATGCAAGAGGATGGTGCTAGTAACGAAACATATACAGCATTAAACGGTTTCTATTCATTTGATAACGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 11 kb fragment | |
| BAC-11kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCGTTATCAAATGAATAATAACCATTAAATGCCATGTATGTATCGTTGTCGTTACCATCTTCGATGTTTGCGT | ||
| BAC-7kb-1 | TACAGGCATTCCGGCAGTTTTAAAATTACTGCCGGAAATGTTTCCTCCTGATTTCTCAAGAGCTGGAACTTGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 7 kb fragment | |
| BAC-7kb-2 | GAAGTACGCTAATGTTTCGTCCACATTTGCGTCAGCAACATCGAGATCTCCGACTTCATAACCAGCACTGATGTTTAAACCGGGTACCGAGCTCGAATTC | ||
| BAC-12kb-1 | TAAAAGAATTGCAAAATGAGTCATATATAATTAAGAAAAATGATCAGGAAGGGGTTATAAAAAGTAATACTGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 12 kb fragment | |
| BAC-12kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTTTAATAGACTCTTTGGCAATGAGTATCGCAGGAACTTCGATTTCTGTTTTTTTATCTTTACTTCTATGTT | ||
| BAC-67kb-1 | TCCTTGATTTAATGTCCAGTTGGGATAGTTATTTACCTAAAATGAAAAAATATAAAAAAGTTATAGGACATGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 67 kb fragment | |
| BAC-67kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACGGATATCACTCTAAAGATATAAGGAATTTTGAAAATATCCCAAAAATCAATAAGTTGATGTTTGAGATAGCT | ||
| BAC-50kb-1 | GAGTTCAGACCCAGCAGATATTCCTCGATATAAAGCAGCCTAAATTACAAAAAGCTTGAAAACTATATAAATGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 50 kb fragment | |
| BAC-50kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTACCCAGGGAGAAGACATATCATGTAAATTAGTCTGAAATCCCATTTCTTCTAATTCATAGCCTATTGGAT |
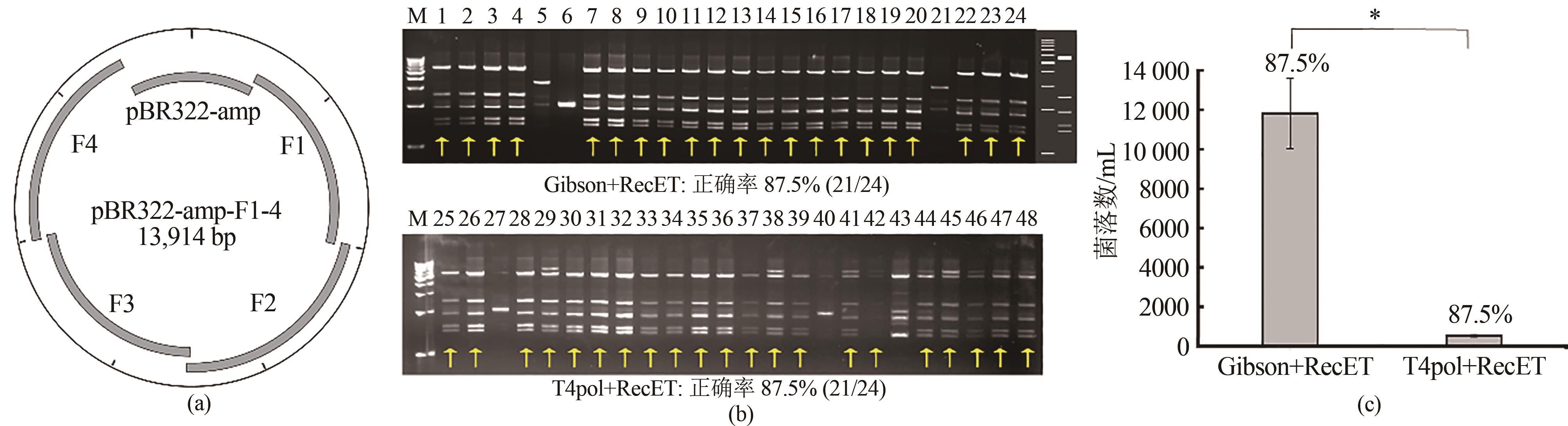
图1 ExoCET技术组装5个DNA片段(a)5个片段组装示意图;(b)组装获得重组质粒的EcoRⅠ酶切图谱,胶图右侧为酶切理论图谱,1~24为Gibson+RecET;25~48为T4pol+RecET;(c)Gibson+RecET和T4pol+RecET五片段组装效率比较。采用单因素方差分析差异。P<0.05认为具有显著性差异(***P < 0.001,**P < 0.01,*P < 0.05)
Fig. 1 Assembly of 5 DNA fragments using ExoCET(a) Schematic diagram for the assembly. (b) EcoRⅠ digestion map for assembled plamids. The right side of the gel imagines is the theoretical map for the restriction digestion. 1-24: Gibson+RecET, and 25-48: T4pol+RecET assembly. (c) Comparison of efficiencies for the assembly. Significance was analyzed by one-way ANOVA, and P < 0.05 was considered statistically significant (***P < 0.001, **P < 0.01, and *P < 0.05)
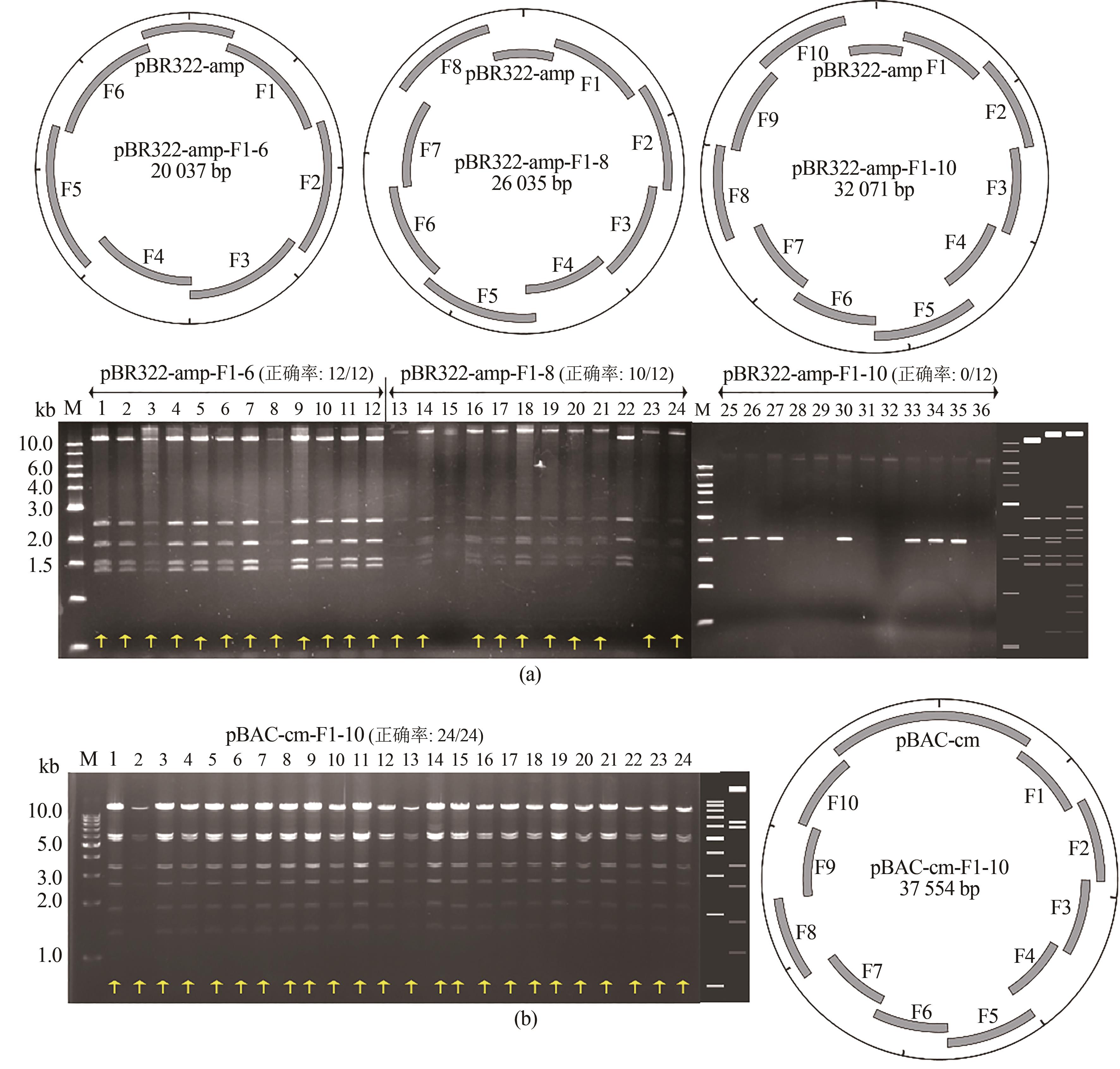
图2 ExoCET技术组装7~11个DNA片段(a)利用pBR322载体组装的示意图和EcoRⅠ酶切鉴定图:1~12为 7个片段组装;13~24为9个片段组装;25~36为11个片段组装;胶图右侧从左到右依次为pBR322-amp-F1-6、pBR322-amp-F1-8、pBR322-amp-F1-10的酶切理论图谱(b)利用BAC载体组装11个片段的示意图和EcoRⅤ酶切鉴定图,胶图右侧为酶切理论图谱
Fig. 2 Assembly of 7-11 fragments using ExoCET(a) Schematic diagrams and EcoRⅠ restriction analysis for the assembly using pBR322 vectors. 1-12: 7-piece assembly, 13-24: 9-piece assembly, and 25-36: 11-piece assembly. The right side of the gel imagine is the theoretical map for the restriction digestion of pBR322-amp-F1-6, pBR322-amp-F1-8 and pBR322-amp-F1-10 from left to right. (b) Schematic diagrams and EcoRⅤ restriction analysis for the 11-piece assembly using the BAC vector. The right side of the gel imagine is the theoretical map for the restriction digestion
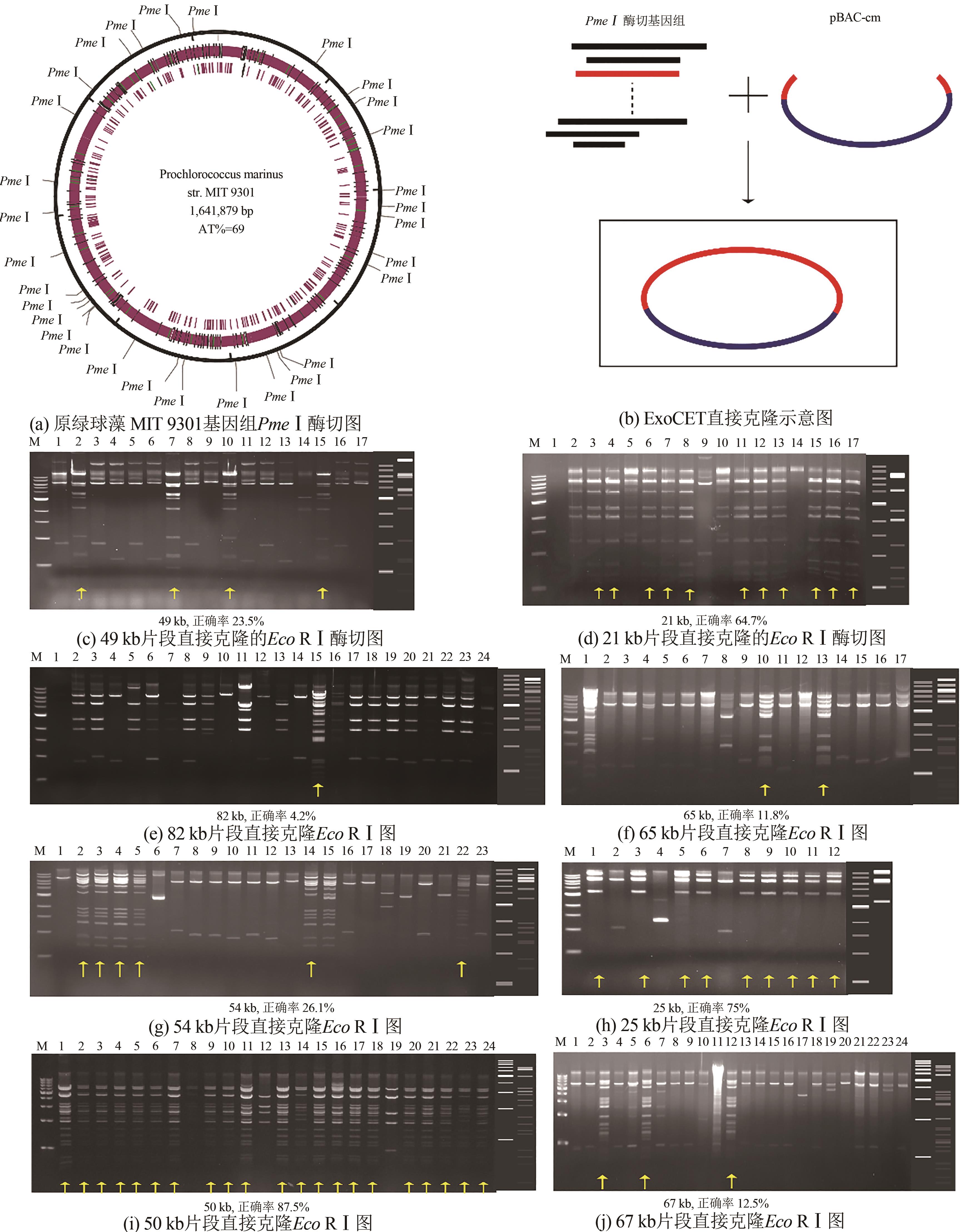
图3 利用ExoCET-BAC策略进行直接基因克隆(a) PmeⅠ restriction map of Prochlorococcus MIT 9301 genome, (b) Strategy for ExoCET direct cloning, and EcoRⅠ restriction map of recombinants from the 49 kb cloning (c), 21 kb cloning (d), 82 kb cloning (e), 65 kb cloning (f), 54 kb cloning (g), 25 kb cloning (h), 50 kb cloning (i), and 67 kb cloning (j), respectively. Yellow arrows indicate correct clones. The right side of the (c)~(j) gel imagines is the theoretical map for the restriction digestion
Fig. 3 ExoCET-BAC strategy for direct gene cloning
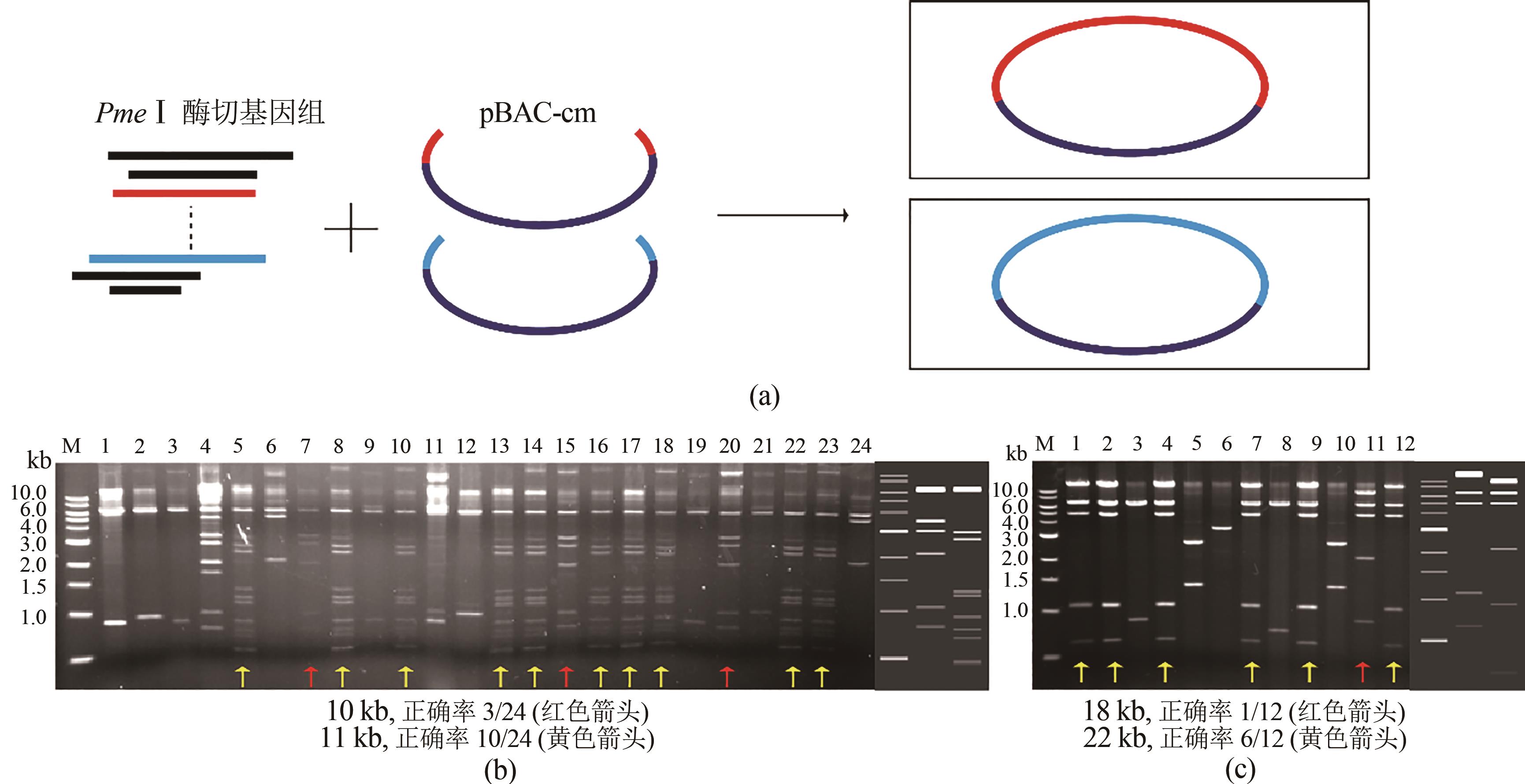
图4 利用ExoCET-BAC策略进行两个片段一步抓取(a)一步抓取两个片段示意图;(b)直接克隆10 kb、11 kb片段EcoRⅠ酶切图(胶图右侧分别为直接克隆10 kb和11 kb基因组大片段的酶切理论图谱);(c)直接克隆18 kb、22 kb片段EcoRⅠ酶切图(胶图右侧从左到右依次为直接克隆22 kb和18 kb基因组大片段的酶切理论图谱)
Fig. 4 ExoCET-BAC strategy for capturing two DNA fragments simultaneously(a) Schematic diagram for the strategy. (b) EcoRⅠrestriction map for the direct cloning of 10 kb and 11 kb fragments. The right side of the gel imagine is the theoretical restriction map for the direct cloning of 10 kb and 11 kb fragments. (c) EcoRⅠ restriction map for the direct cloning of 18 kb and 22 kb fragments. The right side of the gel imagine is the theoretical restriction map for the direct cloning of 18 kb and 22 kb fragments from left to right
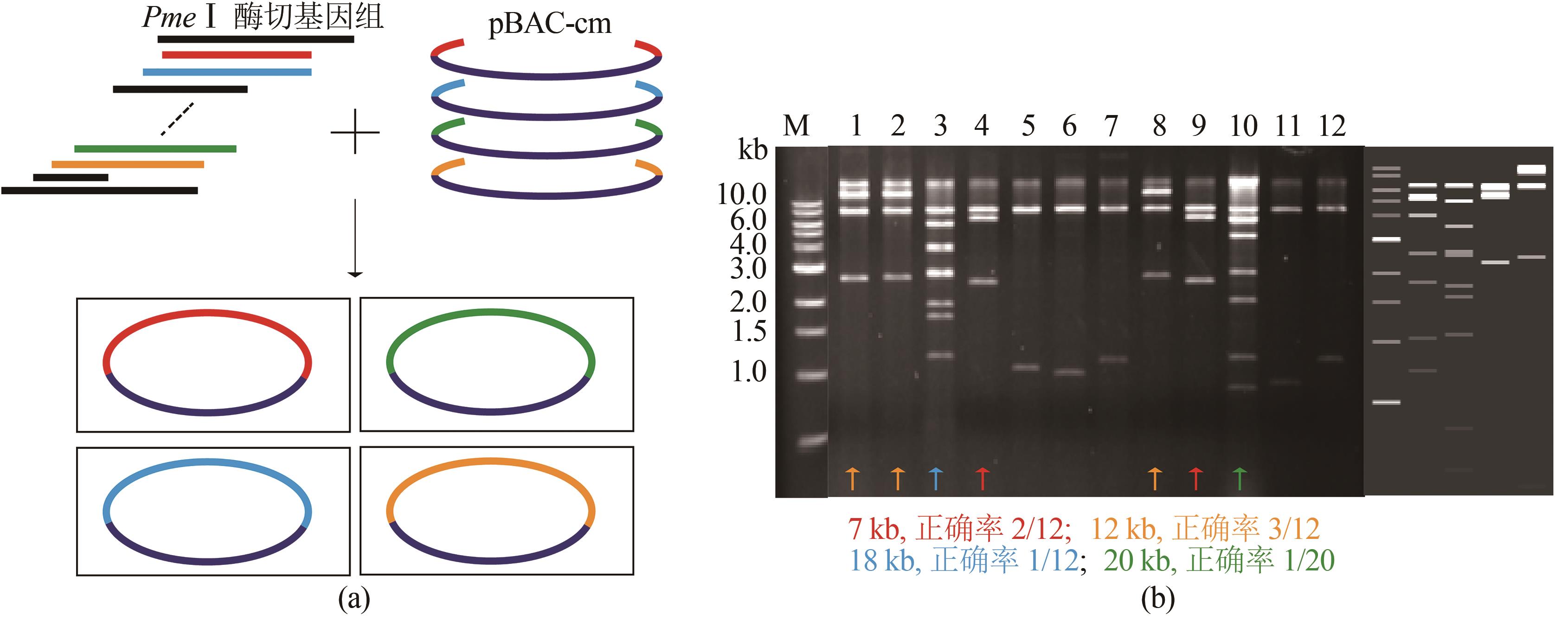
图5 利用ExoCET-BAC策略一步抓取4个片段(a)一步抓取4个片段示意图;(b)直接克隆7 kb、12 kb、18 kb和20 kb的EcoRⅠ酶切结果图(胶图右侧从左到右依次为直接克隆20 kb、18 kb、7 kb和12 kb基因组大片段的酶切理论图谱)
Fig. 5 ExoCET-BAC strategy for capturing four DNA fragments simultaneously(a) Schematic diagram for the strategy. (b) EcoRⅠrestriction map for the direct cloning of 7 kb, 12 kb, 18 kb and 20 kb fragments. The right side of the gel imagine is the theoretical restriction map for the direct cloning of 20 kb、18 kb、7 kb and 12 kb fragments from left to right
| 1 | YAMANAKA K, REYNOLDS K A, KERSTEN R D, et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(5): 1957-1962. |
| 2 | LEE N C O, LARIONOV V, KOUPRINA N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast[J]. Nucleic Acids Research, 2015, 43(8): e55. |
| 3 | FU Jun, BIAN Xiaoying, HU Shenbiao, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting[J]. Nature Biotechnology, 2012, 30(5): 440-446. |
| 4 | JIANG Wenjun, ZHAO Xuejin, GABRIELI T, et al. Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters[J]. Nature Communications, 2015, 6: 8101. |
| 5 | JIANG Wenjun, ZHU Ting F. Targeted isolation and cloning of 100-kb microbial genomic sequences by Cas9-assisted targeting of chromosome segments[J]. Nature Protocols, 2016, 11(5): 960-975. |
| 6 | WANG Hailong, LI Zhen, JIA Ruonan, et al. ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes[J]. Nucleic Acids Research, 2018, 46(5): e28. |
| 7 | TAO Weixin, CHEN Li, ZHAO Chunhua, et al. In vitro packaging mediated one-step targeted cloning of natural product pathway[J]. ACS Synthetic Biology, 2019, 8(9): 1991-1997. |
| 8 | ENGHIAD B, HUANG Chunshuai, GUO Fang, et al. Cas12a-assisted precise targeted cloning using in vivo Cre-lox recombination[J]. Nature Communications, 2021, 12(1): 1171. |
| 9 | GREUNKE C, DUELL E R, D'AGOSTINO P M, et al. Direct Pathway Cloning (DiPaC) to unlock natural product biosynthetic potential[J]. Metabolic Engineering, 2018, 47: 334-345. |
| 10 | D'AGOSTINO P M, GULDER T A M. Direct pathway cloning combined with sequence- and ligation-independent cloning for fast biosynthetic gene cluster refactoring and heterologous expression[J]. ACS Synthetic Biology, 2018, 7(7): 1702-1708. |
| 11 | QUAN Jiayuan, TIAN Jingdong. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries[J]. Nature Protocols, 2011, 6(2): 242-251. |
| 12 | SHAO Zengyi, ZHAO Hua, Zhao Huimin. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways[J]. Nucleic Acids Research, 2009, 37(2): e16. |
| 13 | SHAO Zengyi, ZHAO Huimin. Manipulating natural product biosynthetic pathways via DNA assembler[J]. Current Protocols in Chemical Biology, 2014, 6(2): 65-100. |
| 14 | HIPPEL P H VON, JOHNSON N P, MARCUS A H. Fifty years of DNA “breathing”: reflections on old and new approaches[J]. Biopolymers, 2013, 99(12): 923-954. |
| 15 | GODISKA R, MEAD D, DHODDA V, et al. Linear plasmid vector for cloning of repetitive or unstable sequences in Escherichia coli [J]. Nucleic Acids Research, 2009, 38(6): e88. |
| 16 | HAO Tingting, XIE Zhoujie, WANG Min, et al. An anaerobic bacterium host system for heterologous expression of natural product biosynthetic gene clusters[J]. Nature Communications, 2019, 10: 3665. |
| 17 | ZHANG Youming, BUCHHOLZ F, MUYRERS J P P, et al. A new logic for DNA engineering using recombination in Escherichia coli [J]. Nature Genetics, 1998, 20(2): 123-128. |
| 18 | ZHANG Youming, MUYRERS J P, TESTA G, et al. DNA cloning by homologous recombination in Escherichia coli [J]. Nature Biotechnology, 2000, 18(12): 1314-1317. |
| 19 | WANG Hailong, LI Zhen, JIA Ruonan, et al. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression[J]. Nature Protocols, 2016, 11(7): 1175-1190. |
| 20 | CHAI Yi, SHAN Shiping, WEISSMAN K J, et al. Heterologous expression and genetic engineering of the tubulysin biosynthetic gene cluster using red/ET recombineering and inactivation mutagenesis[J]. Chemistry & Biology, 2012, 19(3): 361-371. |
| 21 | SONG Chaoyi, LUAN Ji, CUI Qingwen, et al. Enhanced heterologous spinosad production from a 79-kb synthetic multi-operon assembly[J]. ACS Synthetic Biology, 2019, 8(1): 137-147. |
| 22 | FU Jun, WENZEL S C, PERLOVA O, et al. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition[J]. Nucleic Acids Research, 2008, 36(17): e113. |
| 23 | ABBASI M N, FU Jun, BIAN Xiaoying, et al. Recombineering for genetic engineering of natural product biosynthetic pathways[J]. Trends in Biotechnology, 2020, 38(7): 715-728. |
| 24 | SONG Chaoyi, LUAN Ji, LI Ruijuan, et al. RedEx: a method for seamless DNA insertion and deletion in large multimodular polyketide synthase gene clusters[J]. Nucleic Acids Research, 2020, 48(22): e130. |
| 25 | WANG Hailong, BIAN Xiaoying, XIA Liqiu, et al. Improved seamless mutagenesis by recombineering using ccdB for counterselection[J]. Nucleic Acids Research, 2013, 42(5): e37. |
| 26 | BIAN Xiaoying, PLAZA A, YAN Fu, et al. Rational and efficient site-directed mutagenesis of adenylation domain alters relative yields of luminmide derivatives in vivo[J]. Biotechnology and Bioengineering, 2015, 112(7): 1343-1353. |
| 27 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019[J]. Journal of Natural Products, 2020, 83(3): 770-803. |
| 28 | HUO Liujie, HUG J J, FU Chengzhang, et al. Heterologous expression of bacterial natural product biosynthetic pathways[J]. Natural Product Reports, 2019, 36(10): 1412-1436. |
| 29 | LI J S, DU Yongle, GU Di, et al. Discovery and biosynthesis of clostyrylpyrones from the obligate anaerobe clostridium roseum[J]. Organic Letters, 2020, 22(21): 8204-8209. |
| 30 | ZHANG Weimin, ZHAO Guanghou, LUO Zhouqing, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355(6329): eaaf3981. |
| 31 | XIE Zexiong, LI Bingzhi, MITCHELL L A, et al. “Perfect” designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 32 | WU Yi, LI Bingzhi, ZHAO Meng, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): eaaf4706. |
| 33 | SHEN Yue, WANG Yun, CHEN Tai, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355(6329): eaaf4791. |
| 34 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 35 | FREDENS J, WANG Kaihang, DE LA TORRE D, et al. Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569(7757): 514-518. |
| [1] | 虞旭昶, 吴辉, 李雷. 文库构建与基因簇靶向筛选驱动的微生物天然产物高效发现[J]. 合成生物学, 2024, 5(3): 492-506. |
| [2] | 奚萌宇, 胡逸灵, 顾玉诚, 戈惠明. 基因组挖掘指导天然药物分子的发现[J]. 合成生物学, 2024, 5(3): 447-473. |
| [3] | 雷茹, 陶慧, 刘天罡. 基因组深度挖掘驱动微生物萜类化合物高效发现[J]. 合成生物学, 2024, 5(3): 507-526. |
| [4] | 宋永相, 张秀凤, 李艳芹, 肖华, 闫岩. 自抗性基因导向的活性天然产物挖掘[J]. 合成生物学, 2024, 5(3): 474-491. |
| [5] | 陈永灿, 司同, 张建志. 自动化合成生物技术在DNA组装与微生物底盘操作中的应用[J]. 合成生物学, 2023, 4(5): 857-876. |
| [6] | 陈青黎, 童贻刚. 工程噬菌体的合成生物学“智造”[J]. 合成生物学, 2023, 4(2): 283-300. |
| [7] | 滕小龙, 史硕博. CRISPR/Cas9系统在基因组编辑中的优化与发展[J]. 合成生物学, 2023, 4(1): 67-85. |
| [8] | 杨毅, 毛雨丰, 杨春贺, 王猛, 廖小平, 马红武. 面向微生物遗传操作的编辑序列设计工具的研究进展[J]. 合成生物学, 2023, 4(1): 30-46. |
| [9] | 潘颖佳, 夏思杨, 董昌, 蔡谨, 连佳长. 基因增变器驱动的酿酒酵母基因组连续进化[J]. 合成生物学, 2023, 4(1): 225-240. |
| [10] | 金交羽, 周佳海. Z-基因组的生物合成奥秘被揭示[J]. 合成生物学, 2022, 3(1): 1-5. |
| [11] | 何博, 付宗恒, 吴毅, 赵广荣. 哺乳动物合成基因组学研究进展[J]. 合成生物学, 2022, 3(1): 78-97. |
| [12] | 杨谦, 程伯涛, 汤志军, 刘文. 基因组挖掘在天然产物发现中的应用和前景[J]. 合成生物学, 2021, 2(5): 697-715. |
| [13] | 周海波, 申琪瑶, 陈汉娜, 王宗杰, 李越中, 张友明, 卞小莹. 利用异源表达挖掘纤维堆囊菌So0157-2的新型天然产物[J]. 合成生物学, 2021, 2(5): 837-849. |
| [14] | 黄小罗, 戴俊彪. 人工DNA合成技术:DNA数据存储的基石[J]. 合成生物学, 2021, 2(3): 335-353. |
| [15] | 汪君仪, 武晓乐, 曹月阳, 李炳志. 基因组设计与合成:从复写到理性设计[J]. 合成生物学, 2021, 2(2): 247-255. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
