合成生物学 ›› 2024, Vol. 5 ›› Issue (5): 1169-1188.DOI: 10.12211/2096-8280.2024-023
一碳生物转化合成有机酸的研究进展
禹伟1,2, 高教琪1,2, 周雍进1,2
- 1.中国科学院大连化学物理研究所,生物技术研究部,辽宁 大连 116023
2.大连市能源生物技术重点实验室,辽宁 大连 116023
-
收稿日期:2024-03-19修回日期:2024-06-04出版日期:2024-10-31发布日期:2024-11-20 -
通讯作者:周雍进 -
作者简介:禹伟 (1992—),男,博士。研究方向为多形汉逊酵母甲醇生物转化与代谢工程。 E-mail:yuweibio@dicp.ac.cn周雍进 (1984—),男,博士,研究员。研究方向为甲醇生物转化与天然产物生物合成。 E-mail:zhouyongjin@dicp.ac.cn -
基金资助:国家重点研发计划(2022YFC2105900);国家自然科学基金(22308351);国家资助博士后研究人员计划(GZB20230727)
Bioconversion of one carbon feedstocks for producing organic acids
YU Wei1,2, GAO Jiaoqi1,2, ZHOU Yongjin1,2
- 1.Division of Biotechnology,Dalian Institute of Chemical Physics,Chinese Academy of Sciences,Dalian 116023,Liaoning,China
2.Dalian Key Laboratory of Energy Biotechnology,Dalian 116023,Liaoning,China
-
Received:2024-03-19Revised:2024-06-04Online:2024-10-31Published:2024-11-20 -
Contact:ZHOU Yongjin
摘要:
有机酸在食品、医药、化工、农业等领域有着广泛的应用。目前有机酸的生产主要以微生物发酵法为主,采用糖类为原料,然而长此以往可能面临“与人争粮”的困境。CO、CO2、甲烷、甲醇和甲酸等含有一个碳原子的物质被称为一碳(one carbon,C1)资源,其来源广泛且价格低廉,有望成为生物制造的替代原料,且C1原料生物转化有助于缓解温室效应、助力“碳中和”目标。本文总结了近年来CO2、甲烷和甲醇生物合成3种重要有机酸(3-羟基丙酸、乳酸、琥珀酸)的研究进展,主要论述了C1生物利用途径、有机酸的生物合成途径以及代谢工程策略,也讨论了C1合成有机酸的挑战及应对措施,并展望了有机酸产业化新路线,尤其是化学催化与生物转化耦合以CO2为原料合成有机酸。本综述对于C1生物炼制以及有机酸产业升级具有一定的参考意义。
中图分类号:
引用本文
禹伟, 高教琪, 周雍进. 一碳生物转化合成有机酸的研究进展[J]. 合成生物学, 2024, 5(5): 1169-1188.
YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids[J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188.
| 产物 | 宿主 | 底物 | 培养条件 | 产量/(g/L) | 得率/% | 生产强度/[g/(L·d)] | 参考文献 |
|---|---|---|---|---|---|---|---|
| 3-HP | 蓝细菌 | CO2 | MM,摇瓶, 50 mL发酵体积 | 0.67 | 2.6 | 0.07 | [ |
| 蓝细菌 | CO2 | MM,摇瓶, 20 mL发酵体积 | 0.84 | 8.0 | 0.14 | [ | |
| 甲基弯菌 | 甲烷 | MM,发酵罐, 50 mL发酵体积 | 0.06 | 2.4 | 0.03 | [ | |
| 扭脱甲基杆菌 | 甲醇 | MM,摇瓶, 50 mL发酵体积 | 0.07 | 2.0 | 0.04 | [ | |
| 扭脱甲基杆菌 | 甲醇 | MM,发酵罐, 1.8 L发酵体积 | 0.86 | 3.1 | 0.21 | [ | |
| 多形汉逊酵母 | 甲醇 | MM,摇瓶, 50 mL发酵体积 | 7.10 | 14.2 | 1.20 | [ | |
| 巴斯德毕赤酵母 | 甲醇 | MM,发酵罐, 300 mL发酵体积 | 48.20 | 23.0 | 3.70 | [ | |
| L-乳酸 | 蓝细菌 | CO2 | MM,摇瓶, 80 mL发酵体积 | 1.00 | N.A. | 0.03 | [ |
| 甲烷氧化菌 | 甲烷 | MM,摇瓶, 2 mL发酵体积 | 0.60 | N.A. | 0.15 | [ | |
| 多形汉逊酵母 | 甲醇 | MM,摇瓶, 50 mL发酵体积 | 3.80 | 8.0 | 0.69 | [ | |
| D-乳酸 | 蓝细菌 | CO2 | MM,发酵罐, 100 mL发酵体积 | 1.30 | N.A. | 0.13 | [ |
| 甲基单胞菌 | 甲烷 | MM,摇瓶, 12.5 mL发酵体积 | 1.20 | 24.5 | 0.20 | [ | |
| 巴斯德毕赤酵母 | 甲醇 | CM,摇瓶, 5 mL发酵体积 | 3.50 | 22.0 | 0.87 | [ | |
| 琥珀酸 | 蓝细菌 | CO2 | MM,发酵罐, 发酵体积未知 | 0.93 | N.A. | 0.19 | [ |
| 蓝细菌 | CO2 | MM,摇瓶, 40 mL发酵体积 | 0.63 | N.A. | 0.32 | [ | |
| 蓝细菌 | CO2 | MM,摇瓶, 发酵体积未知 | 1.80 | N.A. | 0.60 | [ | |
| 蓝细菌 | CO2 | MM,发酵罐, 1 L发酵体积 | 2.50 | N.A. | 0.23 | [ | |
| 甲基单胞菌 | 甲烷 | MM,发酵罐, 3.2 L发酵体积 | 0.20 | 7.9 | 0.04 | [ |
表1 C1原料合成有机酸
Table 1 Bio-production of organic acids from C1 feedstocks
| 产物 | 宿主 | 底物 | 培养条件 | 产量/(g/L) | 得率/% | 生产强度/[g/(L·d)] | 参考文献 |
|---|---|---|---|---|---|---|---|
| 3-HP | 蓝细菌 | CO2 | MM,摇瓶, 50 mL发酵体积 | 0.67 | 2.6 | 0.07 | [ |
| 蓝细菌 | CO2 | MM,摇瓶, 20 mL发酵体积 | 0.84 | 8.0 | 0.14 | [ | |
| 甲基弯菌 | 甲烷 | MM,发酵罐, 50 mL发酵体积 | 0.06 | 2.4 | 0.03 | [ | |
| 扭脱甲基杆菌 | 甲醇 | MM,摇瓶, 50 mL发酵体积 | 0.07 | 2.0 | 0.04 | [ | |
| 扭脱甲基杆菌 | 甲醇 | MM,发酵罐, 1.8 L发酵体积 | 0.86 | 3.1 | 0.21 | [ | |
| 多形汉逊酵母 | 甲醇 | MM,摇瓶, 50 mL发酵体积 | 7.10 | 14.2 | 1.20 | [ | |
| 巴斯德毕赤酵母 | 甲醇 | MM,发酵罐, 300 mL发酵体积 | 48.20 | 23.0 | 3.70 | [ | |
| L-乳酸 | 蓝细菌 | CO2 | MM,摇瓶, 80 mL发酵体积 | 1.00 | N.A. | 0.03 | [ |
| 甲烷氧化菌 | 甲烷 | MM,摇瓶, 2 mL发酵体积 | 0.60 | N.A. | 0.15 | [ | |
| 多形汉逊酵母 | 甲醇 | MM,摇瓶, 50 mL发酵体积 | 3.80 | 8.0 | 0.69 | [ | |
| D-乳酸 | 蓝细菌 | CO2 | MM,发酵罐, 100 mL发酵体积 | 1.30 | N.A. | 0.13 | [ |
| 甲基单胞菌 | 甲烷 | MM,摇瓶, 12.5 mL发酵体积 | 1.20 | 24.5 | 0.20 | [ | |
| 巴斯德毕赤酵母 | 甲醇 | CM,摇瓶, 5 mL发酵体积 | 3.50 | 22.0 | 0.87 | [ | |
| 琥珀酸 | 蓝细菌 | CO2 | MM,发酵罐, 发酵体积未知 | 0.93 | N.A. | 0.19 | [ |
| 蓝细菌 | CO2 | MM,摇瓶, 40 mL发酵体积 | 0.63 | N.A. | 0.32 | [ | |
| 蓝细菌 | CO2 | MM,摇瓶, 发酵体积未知 | 1.80 | N.A. | 0.60 | [ | |
| 蓝细菌 | CO2 | MM,发酵罐, 1 L发酵体积 | 2.50 | N.A. | 0.23 | [ | |
| 甲基单胞菌 | 甲烷 | MM,发酵罐, 3.2 L发酵体积 | 0.20 | 7.9 | 0.04 | [ |
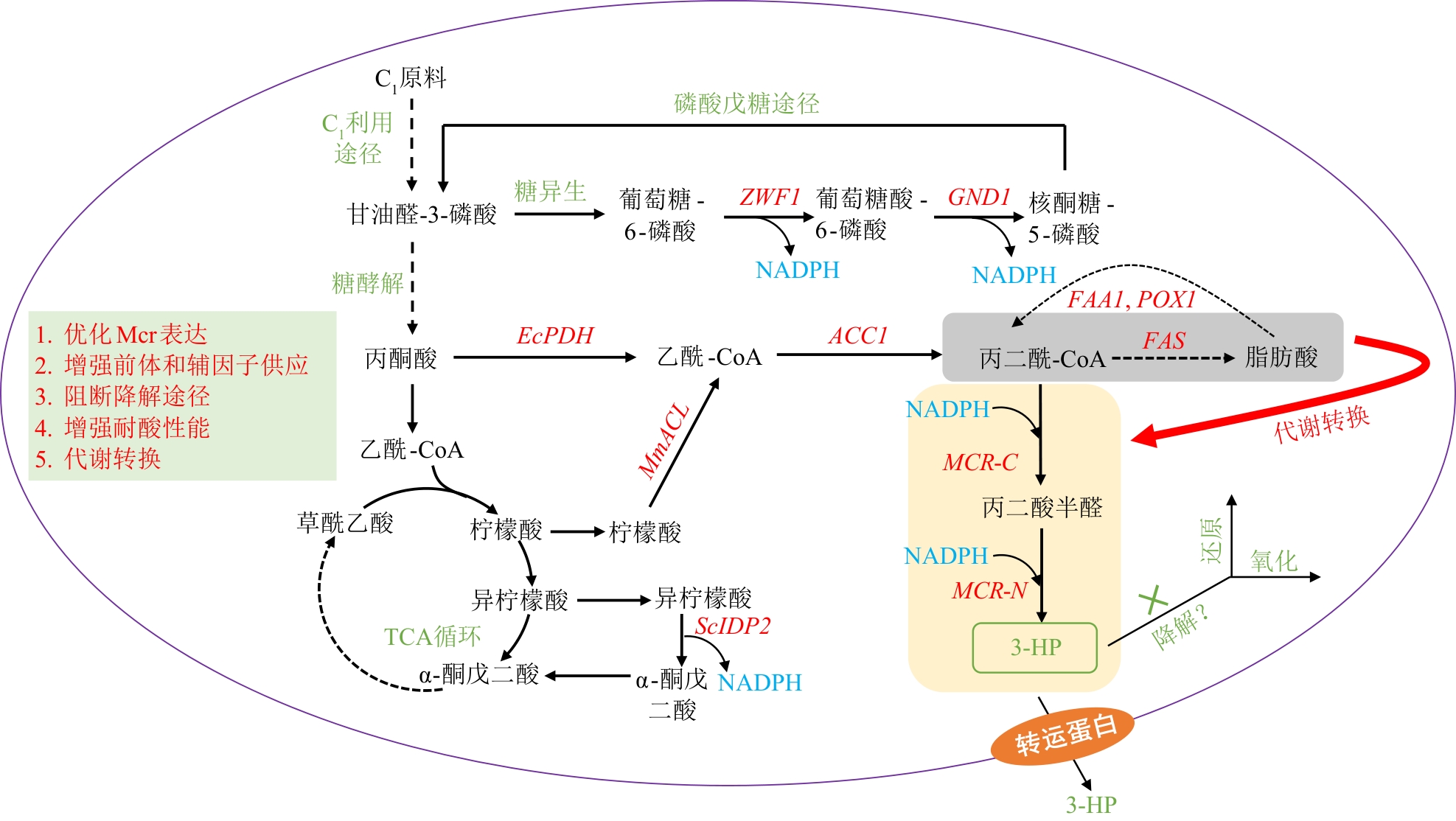
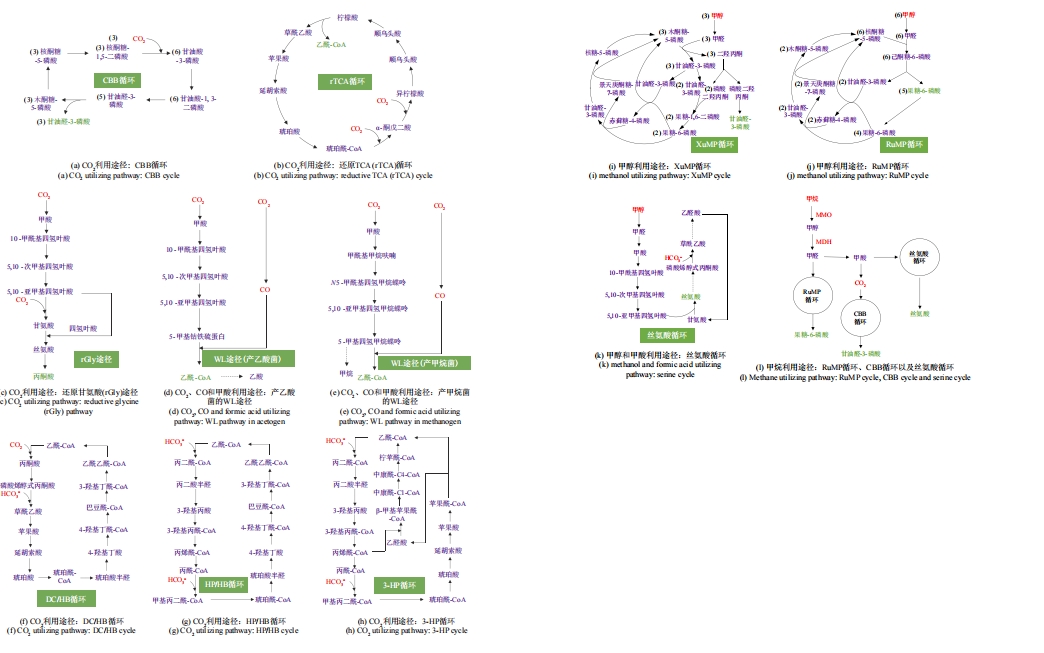
图2 C1原料合成3-HP的代谢途径及其改造策略ZWF1—葡萄糖-6-磷酸脱氢酶基因;GND1—葡萄糖酸-6-磷酸脱氢酶基因;EcPDH—大肠杆菌的丙酮酸脱氢酶系基因;ACC1—乙酰-CoA羧化酶基因;FAA1—脂酰-CoA合成酶基因;POX1—脂酰-CoA氧化酶基因;FAS—脂肪酸合成酶基因;MmACL—小鼠的ATP-柠檬酸裂解酶基因;ScIDP2—酿酒酵母的异柠檬酸脱氢酶基因;MCR-C—丙二酰-CoA还原酶C端基因;MCR-N—丙二酰-CoA还原酶N端基因
Fig. 2 Biosynthetic pathway and engineering strategies for 3-HP production from C1 feedstocksZWF1—Glucose-6-phosphate dehydrogenase gene; GND1—6-Phosphogluconate dehydrogenase gene; EcPDH—Escherichia coli pyruvate dehydrogenase complex gene; ACC1—Acetyl-CoA carboxylase gene; FAA1—Fatty acyl-CoA synthetase gene; POX1—Fatty acyl-CoA oxidase gene; FAS—Fatty acid synthase gene; MmACL—Mouse ATP-citrate lyase gene; ScIDP2—Saccharomyces cerevisiae isocitrate dehydrogenase gene; MCR-C—Malonyl-CoA reductase C-terminal gene; MCR-N—Malonyl-CoA reductase N-terminal gene
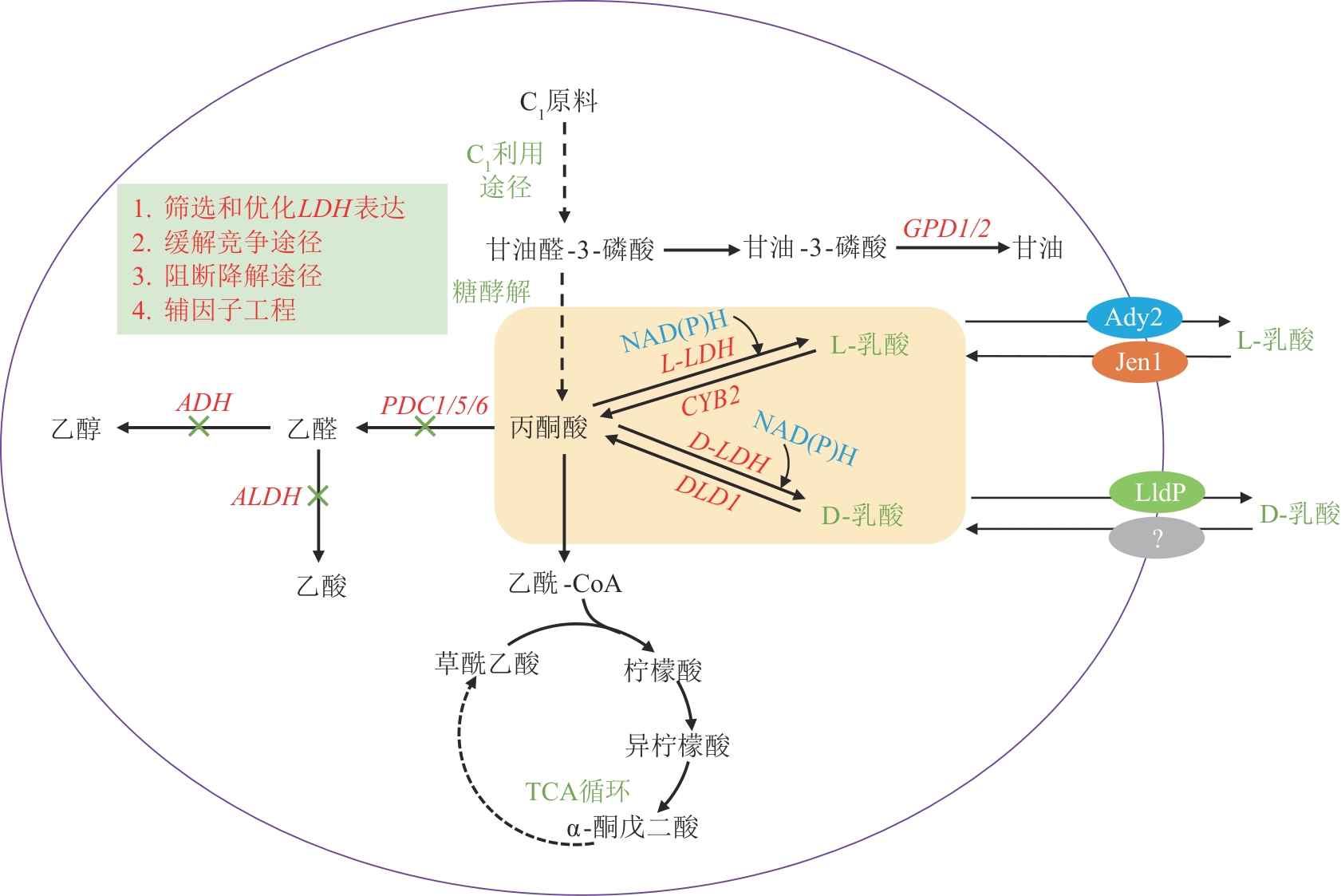
图3 C1原料合成乳酸的代谢途径及其改造策略GPD1/2—甘油-3-磷酸脱氢酶1/2基因;ADH—乙醇脱氢酶基因;PDC1/5/6—丙酮酸脱羧酶1/5/6基因;ALDH—乙醛脱氢酶基因;L-LDH—L-乳酸脱氢酶基因;D-LDH—D-乳酸脱氢酶基因;CYB2—L-乳酸脱氢酶(细胞色素)基因;DLD1—D-乳酸脱氢酶基因;Ady2—乙酸转运蛋白;Jen1—单羧酸/H+同向转运蛋白;LldP—D-乳酸转运蛋白
Fig. 3 Biosynthetic pathway and engineering strategies for lactic acid production from C1 feedstocksGPD1/2—Glycerol-3-phosphate dehydrogenase 1/2 genes; ADH—Alcohol dehydrogenase gene; PDC1/5/6—Pyruvate decarboxylase 1/5/6 genes; ALDH—Aldehyde dehydrogenase gene; L-LDH—l-lactate dehydrogenase gene; D-LDH—D-lactate dehydrogenase gene; CYB2—L-lactate dehydrogenase (cytochrome) gene; DLD1—D-lactate dehydrogenase gene; Ady2—Acetate transporter; Jen1—Monocarboxylate/H+ symporter; LldP—D-lactate transporter
图4 C1原料合成琥珀酸的代谢途径及其改造策略PYC—丙酮酸羧化酶基因;PPC—磷酸烯醇式丙酮酸羧化酶基因;PCK—磷酸烯醇式丙酮酸羧激酶基因;MAE—苹果酸酶基因;MDH—苹果酸脱氢酶基因;FUM—延胡索酸酶基因;FRD—延胡索酸还原酶基因;SDH—琥珀酸脱氢酶基因
Fig. 4 Biosynthetic pathway and engineering strategies for succinic acid production from C1 feedstocksPYC—Pyruvate carboxylase gene; PPC—Phosphoenolpyruvate carboxylase gene; PCK—Phosphoenolpyruvate carboxykinase gene; MAE—Malic enzyme gene; MDH—Malate dehydrogenase gene; FUM—Fumarase gene; FRD—Fumarate reductase gene; SDH—Succinate dehydrogenase gene
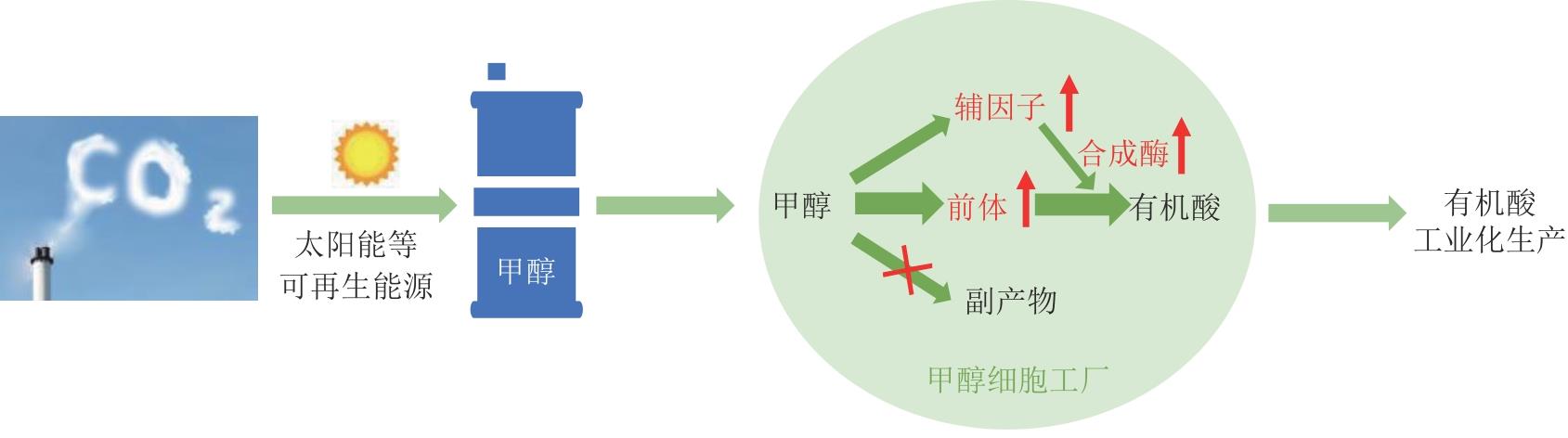
图5 可再生能源催化CO2还原制备甲醇结合甲醇生物转化合成有机酸是未来有机酸生产的潜力路径
Fig. 5 Combination of CO2 reduction and methanol bioconversion is a potential approach for industrial production of organic acids
| 1 | WERPY T, PETERSEN G. Top value added Chemicals from Biomass—VolumeⅠ: results of screening for potential candidates from sugars and synthesis Gas[R/OL]. Pacific Northwest National Laboratory National Renewable Energy Laboratory and Department of Energy. (2004-08-01)[2024-02-01]. . |
| 2 | BOZELL J J, PETERSEN G R. Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy’s “Top 10” revisited[J]. Green Chemistry, 2010, 12(4): 539-554. |
| 3 | 张瑞元, 朱翊凡, 曾杜文, 等. 利用酵母菌生产有机酸的研究进展[J]. 生物工程学报, 2023, 39(6): 2231-2247. |
| ZHANG R Y, ZHU Y F, ZENG D W, et al. Advances on the production of organic acids by yeast[J]. Chinese Journal of Biotechnology, 2023, 39(6): 2231-2247. | |
| 4 | LOMWONGSOPON P, VARRONE C. Contribution of fermentation technology to building blocks for renewable plastics[J]. Fermentation, 2022, 8(2): 47. |
| 5 | 张媛媛, 曾艳, 王钦宏. 合成生物制造进展[J]. 合成生物学, 2021, 2(2): 145-160. |
| ZHANG Y Y, ZENG Y, WANG Q H. Advances in synthetic biomanufacturing[J]. Synthetic Biology Journal, 2021, 2(2): 145-160. | |
| 6 | ZOU L H, OUYANG S P, HU Y L, et al. Efficient lactic acid production from dilute acid-pretreated lignocellulosic biomass by a synthetic consortium of engineered Pseudomonas putida and Bacillus coagulans [J]. Biotechnology for Biofuels, 2021, 14(1): 227. |
| 7 | LI Y, HUGENHOLTZ J, CHEN J, et al. Enhancement of pyruvate production by Torulopsis glabrata using a two-stage oxygen supply control strategy[J]. Applied Microbiology and Biotechnology, 2002, 60(1-2): 101-106. |
| 8 | YU W, CAO X, GAO J Q, et al. Overproduction of 3-hydroxypropionate in a super yeast chassis[J]. Bioresource Technology, 2022, 361: 127690. |
| 9 | ZHOU Y J, KERKHOVEN E J, NIELSEN J. Barriers and opportunities in bio-based production of hydrocarbons[J]. Nature Energy, 2018, 3(11): 925-935. |
| 10 | SANTOS CORREA S, SCHULTZ J, LAUERSEN K J, et al. Natural carbon fixation and advances in synthetic engineering for redesigning and creating new fixation pathways[J]. Journal of Advanced Research, 2023, 47: 75-92. |
| 11 | SARMA S, SHARMA S, RUDAKIYA D, et al. Valorization of microalgae biomass into bioproducts promoting circular bioeconomy: a holistic approach of bioremediation and biorefinery[J]. 3 Biotech, 2021, 11(8): 378. |
| 12 | VEAUDOR T, BLANC-GARIN V, CHENEBAULT C, et al. Recent advances in the photoautotrophic metabolism of cyanobacteria: biotechnological implications[J]. Life, 2020, 10(5): 71. |
| 13 | STEPHENS S, MAHADEVAN R, ALLEN D G. Engineering photosynthetic bioprocesses for sustainable chemical production: a review[J]. Frontiers in Bioengineering and Biotechnology, 2021, 8: 610723. |
| 14 | BAR-EVEN A, NOOR E, LEWIS N E, et al. Design and analysis of synthetic carbon fixation pathways[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(19): 8889-8894. |
| 15 | SCHWANDER T, VON BORZYSKOWSKI L S, BURGENER S, et al. A synthetic pathway for the fixation of carbon dioxide in vitro [J]. Science, 2016, 354(6314): 900-904. |
| 16 | BOUZON M, PERRET A, LOREAU O, et al. A synthetic alternative to canonical one-carbon metabolism[J]. ACS Synthetic Biology, 2017, 6(8): 1520-1533. |
| 17 | LUO S S, DIEHL C, HE H, et al. Construction and modular implementation of the THETA cycle for synthetic CO2 fixation[J]. Nature Catalysis, 2023, 6(12): 1228-1240. |
| 18 | SARWAR A, LEE E Y. Methanol-based biomanufacturing of fuels and chemicals using native and synthetic methylotrophs[J]. Synthetic and Systems Biotechnology, 2023, 8(3): 396-415. |
| 19 | ZHAN C J, LI X W, YANG Y K, et al. Strategies and challenges with the microbial conversion of methanol to high-value chemicals[J]. Biotechnology and Bioengineering, 2021, 118(10): 3655-3668. |
| 20 | ZHAI X X, GAO J Q, LI Y X, et al. Peroxisomal metabolic coupling improves fatty alcohol production from sole methanol in yeast[J]. Proceedings of the National Academy of Sciences of the United States of America, 2023, 120(12): e2220816120. |
| 21 | SEMRAU J D, DISPIRITO A A, YOON S. Methanotrophs and copper[J]. FEMS Microbiology Reviews, 2010, 34(4): 496-531. |
| 22 | KALYUZHNAYA M G, PURI A W, LIDSTROM M E. Metabolic engineering in methanotrophic bacteria[J]. Metabolic Engineering, 2015, 29: 142-152. |
| 23 | BARIK S, PRIETO S, HARRISON S B, et al. Biological production of alcohols from coal through indirect liquefaction[J]. Applied Biochemistry and Biotechnology, 1988, 18(1): 363-378. |
| 24 | KÖPKE M, MIHALCEA C, LIEW F, et al. 2,3-Butanediol production by acetogenic bacteria, an alternative route to chemical synthesis, using industrial waste gas[J]. Applied and Environmental Microbiology, 2011, 77(15): 5467-5475. |
| 25 | FERNÁNDEZ-NAVEIRA Á, ABUBACKAR H N, VEIGA M C, et al. Efficient butanol-ethanol (B-E) production from carbon monoxide fermentation by Clostridium carboxidivorans [J]. Applied Microbiology and Biotechnology, 2016, 100(7): 3361-3370. |
| 26 | LITTY D, KREMP F, MÜLLER V. One substrate, many fates: different ways of methanol utilization in the acetogen Acetobacterium woodii [J]. Environmental Microbiology, 2022, 24(7): 3124-3133. |
| 27 | HEISKANEN H, VIRKAJÄRVI I, VIIKARI L. The effect of syngas composition on the growth and product formation of Butyribacterium methylotrophicum [J]. Enzyme and Microbial Technology, 2007, 41(3): 362-367. |
| 28 | SCHUCHMANN K, MÜLLER V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria[J]. Nature Reviews Microbiology, 2014, 12(12): 809-821. |
| 29 | GOYAL N, ZHOU Z, KARIMI I A. Metabolic processes of Methanococcus maripaludis and potential applications[J]. Microbial Cell Factories, 2016, 15(1): 107. |
| 30 | HEIDELBERG J F, SESHADRI R, HAVEMAN S A, et al. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough[J]. Nature Biotechnology, 2004, 22(5): 554-559. |
| 31 | CROWTHER G J, KOSÁLY G, LIDSTROM M E. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1[J]. Journal of Bacteriology, 2008, 190(14): 5057-5062. |
| 32 | CRAMM R. Genomic view of energy metabolism in Ralstonia eutropha H16[J]. Journal of Molecular Microbiology and Biotechnology, 2009, 16(1-2): 38-52. |
| 33 | GLEIZER S, BEN-NISSAN R, BAR-ON Y M, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2 [J]. Cell, 2019, 179(6): 1255-1263.e12. |
| 34 | GASSLER T, SAUER M, GASSER B, et al. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2 [J]. Nature Biotechnology, 2020, 38(2): 210-216. |
| 35 | TIAN J Z, DENG W, ZHANG Z W, et al. Discovery and remodeling of Vibrio natriegens as a microbial platform for efficient formic acid biorefinery[J]. Nature Communications, 2023, 14(1): 7758. |
| 36 | HENRY C S, BROADBELT L J, HATZIMANIKATIS V. Discovery and analysis of novel metabolic pathways for the biosynthesis of industrial chemicals: 3-hydroxypropanoate[J]. Biotechnology and Bioengineering, 2010, 106(3): 462-473. |
| 37 | KILDEGAARD K R, HALLSTRÖM B M, BLICHER T H, et al. Evolution reveals a glutathione-dependent mechanism of 3-hydroxypropionic acid tolerance[J]. Metabolic Engineering, 2014, 26: 57-66. |
| 38 | SCHWARZ M, KÖPCKE B, WEBER R W, et al. 3-Hydroxypropionic acid as a nematicidal principle in endophytic fungi[J]. Phytochemistry, 2004, 65(15): 2239-2245. |
| 39 | CHEN Y, BAO J C, KIM I K, et al. Coupled incremental precursor and co-factor supply improves 3-hydroxypropionic acid production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2014, 22: 104-109. |
| 40 | LI Y, WANG X, GE X Z, et al. High production of 3-hydroxypropionic acid in Klebsiella pneumoniae by systematic optimization of glycerol metabolism[J]. Scientific Reports, 2016, 6: 26932. |
| 41 | BORODINA I, KILDEGAARD K R, JENSEN N B, et al. Establishing a synthetic pathway for high-level production of 3-hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine[J]. Metabolic Engineering, 2015, 27: 57-64. |
| 42 | JIANG X R, YAN X, YU L P, et al. Hyperproduction of 3-hydroxypropionate by Halomonas bluephagenesis [J]. Nature Communications, 2021, 12(1): 1513. |
| 43 | TONG T, TAO Z Y, CHEN X L, et al. A biosynthesis pathway for 3-hydroxypropionic acid production in genetically engineered Saccharomyces cerevisiae [J]. Green Chemistry, 2021, 23(12): 4502-4509. |
| 44 | ZHAO P, MA C L, XU L D, et al. Exploiting tandem repetitive promoters for high-level production of 3-hydroxypropionic acid[J]. Applied Microbiology and Biotechnology, 2019, 103(10): 4017-4031. |
| 45 | WANG C, REN J, ZHOU L B, et al. An aldolase-catalyzed new metabolic pathway for the assimilation of formaldehyde and methanol to synthesize 2-keto-4-hydroxybutyrate and 1,3-propanediol in Escherichia coli [J]. ACS Synthetic Biology, 2019, 8(11): 2483-2493. |
| 46 | GAO J Q, YU W, LI Y X, et al. Engineering co-utilization of glucose and xylose for chemical overproduction from lignocellulose[J]. Nature Chemical Biology, 2023, 19(12): 1524-1531. |
| 47 | WANG Y P, SUN T, GAO X Y, et al. Biosynthesis of platform chemical 3-hydroxypropionic acid (3-HP) directly from CO2 in cyanobacterium Synechocystis sp. PCC 6803[J]. Metabolic Engineering, 2016, 34: 60-70. |
| 48 | NGUYEN D T N, LEE O K, LIM C, et al. Metabolic engineering of type Ⅱ methanotroph, Methylosinus trichosporium OB3b, for production of 3-hydroxypropionic acid from methane via a malonyl-CoA reductase-dependent pathway[J]. Metabolic Engineering, 2020, 59: 142-150. |
| 49 | WU X Y, CAI P, GAO L H, et al. Efficient bioproduction of 3-hydroxypropionic acid from methanol by a synthetic yeast cell factory[J]. ACS Sustainable Chemistry & Engineering, 2023, 11(16): 6445-6453. |
| 50 | LAN E I, CHUANG D S, SHEN C R, et al. Metabolic engineering of cyanobacteria for photosynthetic 3-hydroxypropionic acid production from CO2 using Synechococcus elongatus PCC 7942[J]. Metabolic Engineering, 2015, 31: 163-170. |
| 51 | YANG Y M, CHEN W J, YANG J, et al. Production of 3-hydroxypropionic acid in engineered Methylobacterium extorquens AM1 and its reassimilation through a reductive route[J]. Microbial Cell Factories, 2017, 16(1): 179. |
| 52 | YUAN X J, CHEN W J, MA Z X, et al. Rewiring the native methanol assimilation metabolism by incorporating the heterologous ribulose monophosphate cycle into Methylorubrum extorquens [J]. Metabolic Engineering, 2021, 64: 95-110. |
| 53 | YU W, GAO J Q, YAO L, et al. Bioconversion of methanol to 3-hydroxypropionate by engineering Ogataea polymorpha [J]. Chinese Journal of Catalysis, 2023, 46: 84-90. |
| 54 | SHABESTARY K, HERNÁNDEZ H P, MIAO R, et al. Cycling between growth and production phases increases cyanobacteria bioproduction of lactate[J]. Metabolic Engineering, 2021, 68: 131-141. |
| 55 | GARG S, CLOMBURG J M, GONZALEZ R. A modular approach for high-flux lactic acid production from methane in an industrial medium using engineered Methylomicrobium buryatense 5GB1[J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(6): 379-391. |
| 56 | WEFELMEIER K, SCHMITZ S, HAUT A M, et al. Engineering the methylotrophic yeast Ogataea polymorpha for lactate production from methanol[J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1223726. |
| 57 | LI C, TAO F, NI J, et al. Enhancing the light-driven production of D-lactate by engineering cyanobacterium using a combinational strategy[J]. Scientific Reports, 2015, 5: 9777. |
| 58 | LEE J K, KIM S J, KIM W S, et al. Efficient production of D-lactate from methane in a lactate-tolerant strain of Methylomonas sp. DH-1 generated by adaptive laboratory evolution[J]. Biotechnology for Biofuels, 2019, 12: 234. |
| 59 | YAMADA R, OGURA K, KIMOTO Y, et al. Toward the construction of a technology platform for chemicals production from methanol: D-lactic acid production from methanol by an engineered yeast Pichia pastoris [J]. World Journal of Microbiology & Biotechnology, 2019, 35(2): 37. |
| 60 | SENGUPTA S, JAISWAL D, SENGUPTA A, et al. Metabolic engineering of a fast-growing Cyanobacterium Synechococcus elongatus PCC 11801 for photoautotrophic production of succinic acid[J]. Biotechnology for Biofuels, 2020, 13: 89. |
| 61 | HUANG C H, SHEN C R, LI H, et al. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942[J]. Microbial Cell Factories, 2016, 15(1): 196. |
| 62 | HASUNUMA T, MATSUDA M, KATO Y, et al. Temperature enhanced succinate production concurrent with increased central metabolism turnover in the cyanobacterium Synechocystis sp. PCC 6803[J]. Metabolic Engineering, 2018, 48: 109-120. |
| 63 | IIJIMA H, WATANABE A, SUKIGARA H, et al. Four-carbon dicarboxylic acid production through the reductive branch of the open cyanobacterial tricarboxylic acid cycle in Synechocystis sp. PCC 6803[J]. Metabolic Engineering, 2021, 65: 88-98. |
| 64 | NGUYEN D T N, LEE O K, HADIYATI S, et al. Metabolic engineering of the typeⅠmethanotroph Methylomonas sp. DH-1 for production of succinate from methane[J]. Metabolic Engineering, 2019, 54: 170-179. |
| 65 | LIU C S, WANG Q, XIAN M, et al. Dissection of malonyl-coenzyme A reductase of Chloroflexus aurantiacus results in enzyme activity improvement[J]. PLoS One, 2013, 8(9): e75554. |
| 66 | LIU C S, DING Y M, ZHANG R B, et al. Functional balance between enzymes in malonyl-CoA pathway for 3-hydroxypropionate biosynthesis[J]. Metabolic Engineering, 2016, 34: 104-111. |
| 67 | ZHOU Y J, BUIJS N A, ZHU Z W, et al. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories[J]. Nature Communications, 2016, 7: 11709. |
| 68 | YU T, ZHOU Y J, HUANG M T, et al. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis[J]. Cell, 2018, 174(6): 1549-1558.e14. |
| 69 | SCHNEIDER K, ASAO M, CARTER M S, et al. Rhodobacter sphaeroides uses a reductive route via propionyl coenzyme A to assimilate 3-hydroxypropionate[J]. Journal of Bacteriology, 2012, 194(2): 225-232. |
| 70 | ZHOU S F, ASHOK S, KO Y, et al. Development of a deletion mutant of Pseudomonas denitrificans that does not degrade 3-hydroxypropionic acid[J]. Applied Microbiology and Biotechnology, 2014, 98(10): 4389-4398. |
| 71 | NGUYEN-VO T P, RYU H, SAUER M, et al. Improvement of 3-hydroxypropionic acid tolerance in Klebsiella pneumoniae by novel transporter YohJK[J]. Bioresource Technology, 2022, 346: 126613. |
| 72 | NGUYEN-VO T P, LIANG Y X, SANKARANARAYANAN M, et al. Development of 3-hydroxypropionic-acid-tolerant strain of Escherichia coli W and role of minor global regulator yieP [J]. Metabolic Engineering, 2019, 53: 48-58. |
| 73 | CHUN A Y, YUNXIAO L, ASHOK S, et al. Elucidation of toxicity of organic acids inhibiting growth of Escherichia coli W[J]. Biotechnology and Bioprocess Engineering, 2014, 19(5): 858-865. |
| 74 | LI J, ZHU K, MIAO L, et al. Simultaneous improvement of limonene production and tolerance in Yarrowia lipolytica through tolerance engineering and evolutionary engineering[J]. ACS Synthetic Biology, 2021, 10(4): 884-896. |
| 75 | YANG M M, AN Y F, ZABED H M, et al. Random mutagenesis of Clostridium butyricum strain and optimization of biosynthesis process for enhanced production of 1,3-propanediol[J]. Bioresource Technology, 2019, 284: 188-196. |
| 76 | ZHANG W, GENG A L. Improved ethanol production by a xylose-fermenting recombinant yeast strain constructed through a modified genome shuffling method[J]. Biotechnology for Biofuels, 2012, 5(1): 46. |
| 77 | ZHU Y, ZHOU C, WANG Y, et al. Transporter engineering for microbial manufacturing[J]. Biotechnology Journal, 2020, 15(9): e1900494. |
| 78 | LIN Z L, ZHANG Y, WANG J Q. Engineering of transcriptional regulators enhances microbial stress tolerance[J]. Biotechnology Advances, 2013, 31(6): 986-991. |
| 79 | CHO J S, KIM G B, EUN H M, et al. Designing microbial cell factories for the production of chemicals[J]. JACS Au, 2022, 2(8): 1781-1799. |
| 80 | NGUYEN-VO T P, KO S, RYU H, et al. Systems evaluation reveals novel transporter YohJK renders 3-hydroxypropionate tolerance in Escherichia coli [J]. Scientific Reports, 2020, 10(1): 19064. |
| 81 | LIU D, HWANG H J, OTOUPAL P B, et al. Engineering Rhodosporidium toruloides for production of 3-hydroxypropionic acid from lignocellulosic hydrolysate[J]. Metabolic Engineering, 2023, 78: 72-83. |
| 82 | VINK E T H, RÁBAGO K R, GLASSNER D A, et al. Applications of life cycle assessment to NatureWorks™ polylactide (PLA) production[J]. Polymer Degradation and Stability, 2003, 80(3): 403-419. |
| 83 | DING X W, RONG J, PAN Z P, et al. De novo multienzyme synthetic pathways for lactic acid production[J]. ACS Catalysis, 2024, 14(7): 4665-4674. |
| 84 | UPADHYAYA B P, DEVEAUX L C, CHRISTOPHER L P. Metabolic engineering as a tool for enhanced lactic acid production[J]. Trends in Biotechnology, 2014, 32(12): 637-644. |
| 85 | HENARD C A, SMITH H, DOWE N, et al. Bioconversion of methane to lactate by an obligate methanotrophic bacterium[J]. Scientific Reports, 2016, 6: 21585. |
| 86 | YU W, GAO J Q, ZHAI X X, et al. Screening neutral sites for metabolic engineering of methylotrophic yeast Ogataea polymorpha [J]. Synthetic and Systems Biotechnology, 2021, 6(2): 63-68. |
| 87 | CAI P, DUAN X P, WU X Y, et al. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris [J]. Nucleic Acids Research, 2021, 49(13): 7791-7805. |
| 88 | WANG M, LUAN G D, LU X F. Systematic identification of a neutral site on chromosome of Synechococcus sp. PCC7002, a promising photosynthetic chassis strain[J]. Journal of Biotechnology, 2019, 295: 37-40. |
| 89 | PORRO D, BIANCHI M M, BRAMBILLA L, et al. Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts[J]. Applied and Environmental Microbiology, 1999, 65(9): 4211-4215. |
| 90 | HIDESE R, MATSUDA M, OSANAI T, et al. Malic enzyme facilitates D-lactate production through increased pyruvate supply during anoxic dark fermentation in Synechocystis sp. PCC 6803[J]. ACS Synthetic Biology, 2020, 9(2): 260-268. |
| 91 | BIANCHI M M, BRAMBILLA L, PROTANI F, et al. Efficient homolactic fermentation by Kluyveromyces lactis strains defective in pyruvate utilization and transformed with the heterologous LDH gene[J]. Applied and Environmental Microbiology, 2001, 67(12): 5621-5625. |
| 92 | ANGERMAYR S A, VAN DER WOUDE A D, CORREDDU D, et al. Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803[J]. Biotechnology for Biofuels, 2014, 7: 99. |
| 93 | BAEK S H, KWON E Y, KIM Y H, et al. Metabolic engineering and adaptive evolution for efficient production of D-lactic acid in Saccharomyces cerevisiae [J]. Applied Microbiology and Biotechnology, 2016, 100(6): 2737-2748. |
| 94 | PACHECO A, TALAIA G, SÁ-PESSOA J, et al. Lactic acid production in Saccharomyces cerevisiae is modulated by expression of the monocarboxylate transporters Jen1 and Ady2[J]. FEMS Yeast Research, 2012, 12(3): 375-381. |
| 95 | WAKAMATSU M, TOMITAKA M, TANI T, et al. Improvement of ethanol production from D-lactic acid by constitutive expression of lactate transporter Jen1p in Saccharomyces cerevisiae [J]. Bioscience, Biotechnology, and Biochemistry, 2013, 77(5): 1114-1116. |
| 96 | GUIARD B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2)[J]. The EMBO Journal, 1985, 4(12): 3265-3272. |
| 97 | MOURIER A, VALLORTIGARA J, YOBOUE E D, et al. Kinetic activation of yeast mitochondrial D-lactate dehydrogenase by carboxylic acids[J]. Biochimica et Biophysica Acta (BBA) - Bioenergetics, 2008, 1777(10): 1283-1288. |
| 98 | BAUMSCHABL M, ATA Ö, MITIC B M, et al. Conversion of CO2 into organic acids by engineered autotrophic yeast[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(47): e2211827119. |
| 99 | TONG T, CHEN X L, HU G P, et al. Engineering microbial metabolic energy homeostasis for improved bioproduction[J]. Biotechnology Advances, 2021, 53: 107841. |
| 100 | LEE J Y, KANG C D, LEE S H, et al. Engineering cellular redox balance in Saccharomyces cerevisiae for improved production of L-lactic acid[J]. Biotechnology and Bioengineering, 2015, 112(4): 751-758. |
| 101 | KOMATI REDDY G, LINDNER S N, WENDISCH V F. Metabolic engineering of an ATP-neutral Embden-Meyerhof-Parnas pathway in Corynebacterium glutamicum: growth restoration by an adaptive point mutation in NADH dehydrogenase[J]. Applied and Environmental Microbiology, 2015, 81(6): 1996-2005. |
| 102 | QI H S, LI S S, ZHAO S M, et al. Model-driven redox pathway manipulation for improved isobutanol production in Bacillus subtilis complemented with experimental validation and metabolic profiling analysis[J]. PLoS One, 2014, 9(4): e93815. |
| 103 | MULLINEAUX C W. Electron transport and light-harvesting switches in cyanobacteria[J]. Frontiers in Plant Science, 2014, 5: 7. |
| 104 | LIU X T, ZHAO G, SUN S J, et al. Biosynthetic pathway and metabolic engineering of succinic acid[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 843887. |
| 105 | LIU Y P, ZHENG P, SUN Z H, et al. Economical succinic acid production from cane molasses by Actinobacillus succinogenes [J]. Bioresource Technology, 2008, 99(6): 1736-1742. |
| 106 | LEE P C, LEE S Y, HONG S H, et al. Batch and continuous cultures of Mannheimia succiniciproducens MBEL55E for the production of succinic acid from whey and corn steep liquor[J]. Bioprocess and Biosystems Engineering, 2003, 26(1): 63-67. |
| 107 | MEYNIAL-SALLES I, DOROTYN S, SOUCAILLE P. A new process for the continuous production of succinic acid from glucose at high yield, titer, and productivity[J]. Biotechnology and Bioengineering, 2008, 99(1): 129-135. |
| 108 | KUHNERT P, SCHOLTEN E, HAEFNER S, et al. Basfia succiniciproducens gen. nov., sp. nov., a new member of the family Pasteurellaceae isolated from bovine rumen[J]. International Journal of Systematic and Evolutionary Microbiology, 2010, 60(Pt 1): 44-50. |
| 109 | LEE S J, LEE D Y, KIM T Y, et al. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation[J]. Applied and Environmental Microbiology, 2005, 71(12): 7880-7887. |
| 110 | LITSANOV B, BROCKER M, BOTT M. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate[J]. Applied and Environmental Microbiology, 2012, 78(9): 3325-3337. |
| 111 | CUI Z Y, GAO C J, LI J J, et al. Engineering of unconventional yeast Yarrowia lipolytica for efficient succinic acid production from glycerol at low pH[J]. Metabolic Engineering, 2017, 42: 126-133. |
| 112 | LAI M J, TSAI J C, LAN E I. CRISPRi-enhanced direct photosynthetic conversion of carbon dioxide to succinic acid by metabolically engineered cyanobacteria[J]. Bioresource Technology, 2022, 366: 128131. |
| 113 | HASUNUMA T, MATSUDA M, KONDO A. Improved sugar-free succinate production by Synechocystis sp. PCC 6803 following identification of the limiting steps in glycogen catabolism[J]. Metabolic Engineering Communications, 2016, 3: 130-141. |
| 114 | LAN E I, WEI C T. Metabolic engineering of cyanobacteria for the photosynthetic production of succinate[J]. Metabolic Engineering, 2016, 38: 483-493. |
| 115 | LU S Y, EITEMAN M A, ALTMAN E. Effect of CO2 on succinate production in dual-phase Escherichia coli fermentations[J]. Journal of Biotechnology, 2009, 143(3): 213-223. |
| 116 | COTELESAGE J J H, PUTTICK J, GOLDIE H, et al. How does an enzyme recognize CO2?[J]. The International Journal of Biochemistry & Cell Biology, 2007, 39(6): 1204-1210. |
| 117 | XIAO M Y, ZHU X N, BI C H, et al. Improving succinate productivity by engineering a cyanobacterial CO2 concentrating system (CCM) in Escherichia coli [J]. Biotechnology Journal, 2017, 12(9): 1700199. |
| 118 | PRICE G D, WOODGER F J, BADGER M R, et al. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(52): 18228-18233. |
| 119 | SHIBATA M, KATOH H, SONODA M, et al. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis[J]. Journal of Biological Chemistry, 2002, 277(21): 18658-18664. |
| 120 | ZHU L W, ZHANG L, WEI L N, et al. Collaborative regulation of CO2 transport and fixation during succinate production in Escherichia coli [J]. Scientific Reports, 2015, 5: 17321. |
| 121 | DURALL C, KUKIL K, HAWKES J A, et al. Production of succinate by engineered strains of Synechocystis PCC 6803 overexpressing phosphoenolpyruvate carboxylase and a glyoxylate shunt[J]. Microbial Cell Factories, 2021, 20(1): 39. |
| 122 | TAPSCOTT T, GUARNIERI M T, HENARD C A. Development of a CRISPR/Cas9 system for Methylococcus capsulatus in vivo gene editing[J]. Applied and Environmental Microbiology, 2019, 85(11): e00340-19. |
| 123 | MO X H, ZHANG H, WANG T M, et al. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis[J]. Applied Microbiology and Biotechnology, 2020, 104(10): 4515-4532. |
| 124 | SCHULTENKÄMPER K, BRITO L F, LÓPEZ M G, et al. Establishment and application of CRISPR interference to affect sporulation, hydrogen peroxide detoxification, and mannitol catabolism in the methylotrophic thermophile Bacillus methanolicus [J]. Applied Microbiology and Biotechnology, 2019, 103(14): 5879-5889. |
| 125 | GAO J Q, GAO N, ZHAI X X, et al. Recombination machinery engineering for precise genome editing in methylotrophic yeast Ogataea polymorpha [J]. iScience, 2021, 24(3): 102168. |
| 126 | ZHAI X X, JI L L, GAO J Q, et al. Characterizing methanol metabolism-related promoters for metabolic engineering of Ogataea polymorpha [J]. Applied Microbiology and Biotechnology, 2021, 105(23): 8761-8769. |
| 127 | NIELSEN J, KEASLING J D. Engineering cellular metabolism[J]. Cell, 2016, 164(6): 1185-1197. |
| 128 | LIEBAL U W, FABRY B A, RAVIKRISHNAN A, et al. Genome-scale model reconstruction of the methylotrophic yeast Ogataea polymorpha [J]. BMC Biotechnology, 2021, 21(1): 23. |
| 129 | KING Z A, LU J, DRÄGER A, et al. BiGG Models: a platform for integrating, standardizing and sharing genome-scale models[J]. Nucleic Acids Research, 2016, 44(D1): D515-D522. |
| 130 | SHIH C F, ZHANG T, LI J H, et al. Powering the future with liquid sunshine[J]. Joule, 2018, 2(10): 1925-1949. |
| [1] | 郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44. |
| [2] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [3] | 赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318. |
| [4] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [5] | 陈锡玮, 张华然, 邹懿. 真菌源非核糖体肽类药物生物合成及代谢工程[J]. 合成生物学, 2024, 5(3): 571-592. |
| [6] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [7] | 惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671. |
| [8] | 赵静宇, 张健, 祁庆生, 王倩. 基于细菌双组分系统的生物传感器的研究进展[J]. 合成生物学, 2024, 5(1): 38-52. |
| [9] | 孙绘梨, 崔金玉, 栾国栋, 吕雪峰. 面向高效光驱固碳产醇的蓝细菌合成生物技术研究进展[J]. 合成生物学, 2023, 4(6): 1161-1177. |
| [10] | 晏雄鹰, 王振, 娄吉芸, 张皓瑜, 黄星宇, 王霞, 杨世辉. 生物燃料高效生产微生物细胞工厂构建研究进展[J]. 合成生物学, 2023, 4(6): 1082-1121. |
| [11] | 程真真, 张健, 高聪, 刘立明, 陈修来. 代谢工程改造微生物利用甲酸研究进展[J]. 合成生物学, 2023, 4(4): 756-778. |
| [12] | 刘家宇, 杨智晗, 杨蕾, 朱丽英, 朱政明, 江凌. 合成生物技术驱动酪丁酸梭菌细胞工厂开发的研究进展[J]. 合成生物学, 2022, 3(6): 1174-1200. |
| [13] | 郭姝媛, 吴良焕, 刘香健, 王博, 于涛. 微生物中一碳代谢网络构建的进展与挑战[J]. 合成生物学, 2022, 3(1): 116-137. |
| [14] | 陈久洲, 王钰, 蒲伟, 郑平, 孙际宾. 5-氨基乙酰丙酸生物合成技术的发展及展望[J]. 合成生物学, 2021, 2(6): 1000-1016. |
| [15] | 汪庆卓, 宋萍, 黄和. 合成生物技术驱动天然的真核油脂细胞工厂开发[J]. 合成生物学, 2021, 2(6): 920-941. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||