合成生物学 ›› 2021, Vol. 2 ›› Issue (2): 256-273.DOI: 10.12211/2096-8280.2020-073
工业丝状真菌基因组编辑技术研究进展
刘倩1,2, 李金根1,2, 张晨阳1,2,3, 李芳雅1,2, 田朝光1,2,3
- 1.中国科学院天津工业生物技术研究所,系统微生物工程重点实验室,天津 300308
2.国家合成生物技术创新中心,天津 300308
3.中国科学院大学,北京 100049
-
收稿日期:2020-07-09修回日期:2021-03-15出版日期:2021-04-30发布日期:2021-04-30 -
通讯作者:田朝光 -
作者简介:刘倩 (1983─),女,博士,副研究员,研究方向为工业真菌遗传改造。E-mail:liu_q1@tib.cas.cn
田朝光(1973─),男,博士,研究员,研究方向为真菌合成生物学。E-mail:tian_cg@tib.cas.cn -
基金资助:国家重点研发计划“合成生物学”重点专项(2018FYA0900500);天津市合成生物技术创新能力提升行动项目(TSBICIP-KJGG-006);国家自然科学基金项目(31972878);中国科学院青年创新促进会项目(2019180)
Research progress of genome editing technologies for industrial filamentous fungi
LIU Qian1,2, LI Jingen1,2, ZHANG Chenyang1,2,3, LI Fangya1,2, TIAN Chaoguang1,2,3
- 1.Key Laboratory of Systems Microbial Biotechnology,Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,Tianjin 300308,China
2.National Technology Innovation Center of Synthetic Biology,Tianjin 300308,China
3.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2020-07-09Revised:2021-03-15Online:2021-04-30Published:2021-04-30 -
Contact:TIAN Chaoguang
摘要:
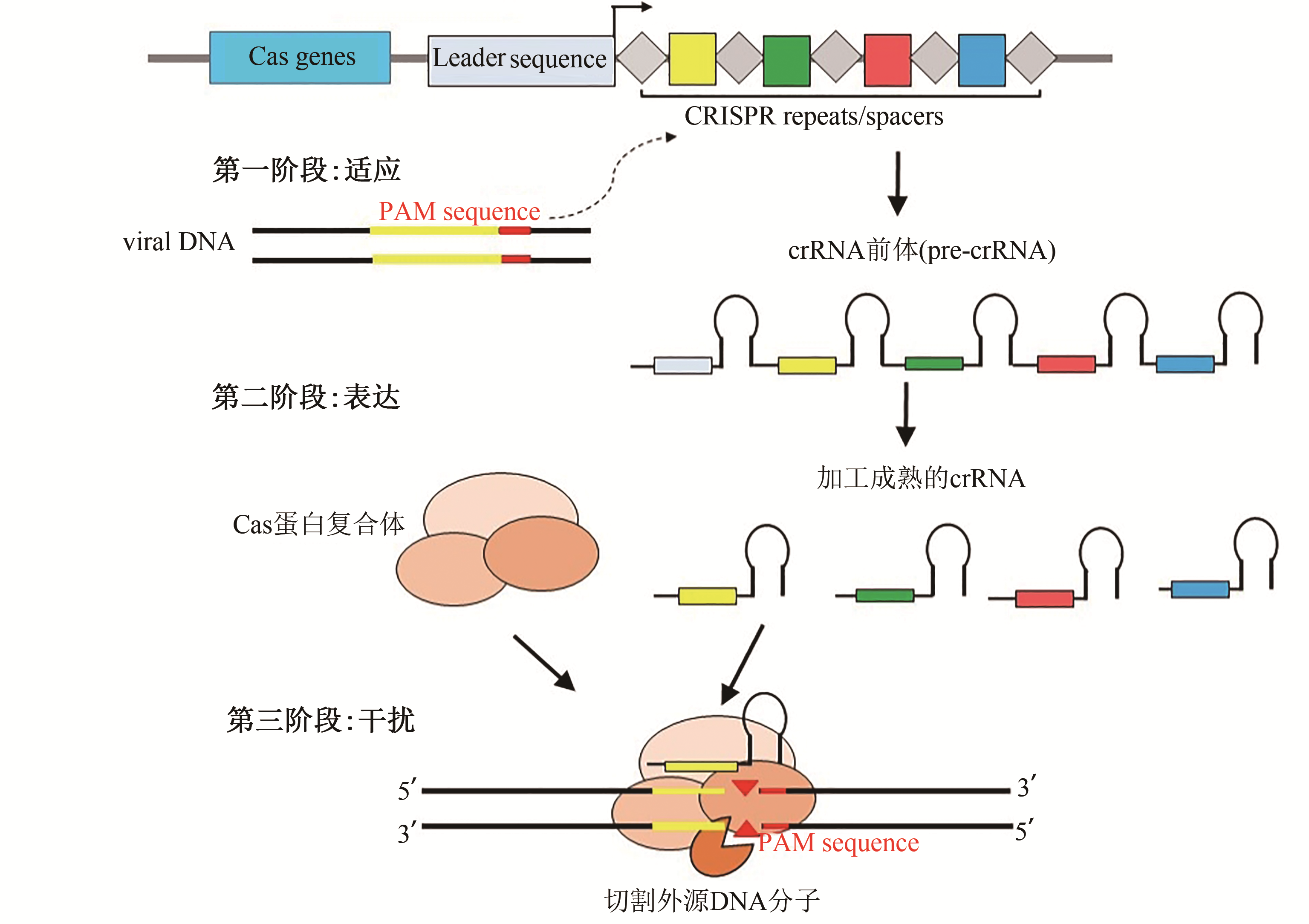
丝状真菌(filamentous fungi)是广泛存在于自然界中的一类多细胞真核微生物,是酶制剂、有机酸、抗生素的核心生产体系,在生物技术中发挥着非常重要的作用。由于丝状真菌的生长发育较为复杂,其遗传体系和基因组编辑技术发展相对较慢,妨碍了丝状真菌基础研究和开发利用的快速发展。近年来,核酸酶介导的基因组编辑技术已经发展成为一种功能强大的基因组编辑工具,在生物技术领域的应用得到广泛关注,其中RNA介导的CRISPR系统(clustered regularly interspaced short palindromic repeats)已经成为新一代基因组编辑技术体系。文章将对目前使用最为广泛的3种基因编辑系统,包括CRISPR-Cas技术的发展历程进行概述,对CRISPR-Cas技术在丝状真菌基因组编辑中的研发进行系统介绍,包括在工业丝状真菌中的研发应用,例如鉴定次级代谢产物的关键基因并提高次级代谢物的生产能力、将外源基因定点整合到基因组上从而提高异源蛋白的表达、遗传重构蛋白分泌途径提高工业酶的产量等,最后对CRISPR-Cas基因组编辑技术及其衍生系统在丝状真菌的真菌基因功能研究、代谢途径重构、精确表达调控、蛋白定向进化以及高性能底盘构建等方面进行了展望。
中图分类号:
引用本文
刘倩, 李金根, 张晨阳, 李芳雅, 田朝光. 工业丝状真菌基因组编辑技术研究进展[J]. 合成生物学, 2021, 2(2): 256-273.
LIU Qian, LI Jingen, ZHANG Chenyang, LI Fangya, TIAN Chaoguang. Research progress of genome editing technologies for industrial filamentous fungi[J]. Synthetic Biology Journal, 2021, 2(2): 256-273.

图1 三种核酸酶介导的基因组编辑模式示意图
Fig. 1 Genome editing using sequence-specific double-stranded breaks by the programmable nucleases including ZFNs, TALENs and CRISPR
| 丝状真菌 | Cas 类型 | Cas启动子 | sgRNA启动子 | 靶标基因 | 编辑效率 | 参考文献 |
|---|---|---|---|---|---|---|
| 黑曲霉 | Cas9 | P tef1 | P gpdA | albA | 高效 | [ |
| P tef1 | Af_U6-1p | albA | 高效 | [ | ||
| P tef1 | PanU6 | albA | 79% | [ | ||
| P gpdA | 5S rRNA | alba, fum5, fum1 | 高达100% | [ | ||
| P tef1 | AoU6, AfU6 | fwnA, amyA, glaA | 单基因高达80% | [ | ||
| P tef1, P gpdA, P glaA | Anp | pyrG, moc, laeA | 高达97.2% | [ | ||
| LbCpf1 | P tef1 | Af_U3p | albA | 80% | [ | |
| 米曲霉 | Cas9 | P amyB | 米曲霉U6p | wA, pyrG, yA | 10%~100% | [ |
| P Aotef1, AMA1-自主复制质粒 | PU6p,AMA1-自主复制质粒 | wA, pyrG, yA | 55.6%~100% | [ | ||
| 里氏木霉 | Cas9 | P pdc, P cbh1 | 体外转录合成 | ura5, lae1, vib1, clr2 | 单基因高达93%~100%;多基因同时编辑效率4.2%~45% | [ |
| 体外转录合成 | 体外转录合成 | cbh-1 | 40% | [ | ||
| P pdc | 里氏木霉PU61, PU62 | ura5 | 高效 | [ | ||
| 体外转录合成 | 体外转录合成 | Trura5,Trlae1,Trcbh1,Trcbh2, Treg1 | 56.52%~100% | [ | ||
| 产黄青霉 | Cas9 | P xyl 或体外 转录合成 | U6, 145 tRNA, Utp25或体外转录合成 | pks17, roqA, lovF,pcbAB, penDE, hcpA | 高达60%~100% | [ |
| 嗜热毁丝霉 | Cas9 | P tef1 | 嗜热毁丝霉MtU6p | amd S, cre1, res1, alp1, gh1-1, rca1, hcr1, ap3, prk6 | 单基因高达95%;双、三、四基因同时编辑效率22%~70% | [ |
| SpCas9, FnCpf1, AsCpf1 | P tef1 或体外 转录合成 | U6p或体外 转录合成 | pks4.2, alp1, snc1, ptf1 | 单基因高达100%;多基因同时编辑时单基因效率5%~100% | [ | |
| AsCas12a | P tef1 | MtU6p | cre1, res1, alp1, gh1-1, rca1,neo, bar, hcr1, ap3, prk6 | 单基因高达90%;三、四基因同时编辑效率22%~41% | [ | |
| 棉阿舒囊霉 | SpCas9 | 酿酒酵母P tef1 | P snr52 | ade2, A754, fmp27 | 36%~80% | [ |
| LbCpf1 | P tsa1 | P snr52 | his3, ade2, trp1, leu2, ura3 | 多基因编辑效率10.4%~77.2% | [ |
表1 工业丝状真菌中CRISPR/Cas系统的技术应用
Tab. 1 Applycations of CRISPR-Cas systems in industrial filamentous fungi
| 丝状真菌 | Cas 类型 | Cas启动子 | sgRNA启动子 | 靶标基因 | 编辑效率 | 参考文献 |
|---|---|---|---|---|---|---|
| 黑曲霉 | Cas9 | P tef1 | P gpdA | albA | 高效 | [ |
| P tef1 | Af_U6-1p | albA | 高效 | [ | ||
| P tef1 | PanU6 | albA | 79% | [ | ||
| P gpdA | 5S rRNA | alba, fum5, fum1 | 高达100% | [ | ||
| P tef1 | AoU6, AfU6 | fwnA, amyA, glaA | 单基因高达80% | [ | ||
| P tef1, P gpdA, P glaA | Anp | pyrG, moc, laeA | 高达97.2% | [ | ||
| LbCpf1 | P tef1 | Af_U3p | albA | 80% | [ | |
| 米曲霉 | Cas9 | P amyB | 米曲霉U6p | wA, pyrG, yA | 10%~100% | [ |
| P Aotef1, AMA1-自主复制质粒 | PU6p,AMA1-自主复制质粒 | wA, pyrG, yA | 55.6%~100% | [ | ||
| 里氏木霉 | Cas9 | P pdc, P cbh1 | 体外转录合成 | ura5, lae1, vib1, clr2 | 单基因高达93%~100%;多基因同时编辑效率4.2%~45% | [ |
| 体外转录合成 | 体外转录合成 | cbh-1 | 40% | [ | ||
| P pdc | 里氏木霉PU61, PU62 | ura5 | 高效 | [ | ||
| 体外转录合成 | 体外转录合成 | Trura5,Trlae1,Trcbh1,Trcbh2, Treg1 | 56.52%~100% | [ | ||
| 产黄青霉 | Cas9 | P xyl 或体外 转录合成 | U6, 145 tRNA, Utp25或体外转录合成 | pks17, roqA, lovF,pcbAB, penDE, hcpA | 高达60%~100% | [ |
| 嗜热毁丝霉 | Cas9 | P tef1 | 嗜热毁丝霉MtU6p | amd S, cre1, res1, alp1, gh1-1, rca1, hcr1, ap3, prk6 | 单基因高达95%;双、三、四基因同时编辑效率22%~70% | [ |
| SpCas9, FnCpf1, AsCpf1 | P tef1 或体外 转录合成 | U6p或体外 转录合成 | pks4.2, alp1, snc1, ptf1 | 单基因高达100%;多基因同时编辑时单基因效率5%~100% | [ | |
| AsCas12a | P tef1 | MtU6p | cre1, res1, alp1, gh1-1, rca1,neo, bar, hcr1, ap3, prk6 | 单基因高达90%;三、四基因同时编辑效率22%~41% | [ | |
| 棉阿舒囊霉 | SpCas9 | 酿酒酵母P tef1 | P snr52 | ade2, A754, fmp27 | 36%~80% | [ |
| LbCpf1 | P tsa1 | P snr52 | his3, ade2, trp1, leu2, ura3 | 多基因编辑效率10.4%~77.2% | [ |
| 88 | QIN H, XIAO H, ZOU G, et al. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species[J]. Process Biochemistry, 2017, 56: 57-61. |
| 89 | SUGANO S S, SUZUKI H, SHIMOKITA E, et al. Genome editing in the mushroom-forming basidiomycete Coprinopsis cinerea, optimized by a high-throughput transformation system[J]. Scientific Reports, 2017, 7(1): 1260. |
| 90 | CHEN B X, WEI T, YE Z W, et al. Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris [J]. Frontiers in Microbiology, 2018, 9: 1157. |
| 91 | DENG H, GAO R, LIAO X, et al. Genome editing in Shiraia bambusicola using CRISPR-Cas9 system[J]. Journal of Biotechnology, 2017, 259: 228-234. |
| 92 | HU J H, MILLER S M, GEURTS M H, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity[J]. Nature, 2018, 556(7699): 57-63. |
| 93 | NISHIMASU H, SHI X, ISHIGURO S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space[J]. Science, 2018, 361(6408): 1259-1262. |
| 94 | MILLER S M, WANG T, RANDOLPH P B, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs[J]. Nature Biotechnology, 2020, 38(4): 471-481. |
| 95 | WALTON R T, CHRISTIE K A, WHITTAKER M N, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296. |
| 96 | TENG F, CUI T, FENG G, et al. Repurposing CRISPR-Cas12b for mammalian genome engineering[J]. Cell Discovery, 2018, 4: 63. |
| 97 | STRECKER J, JONES S, KOOPAL B, et al. Engineering of CRISPR-Cas12b for human genome editing[J]. Nature Communications 2019, 10(1): 21. |
| 98 | MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants[J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. |
| 99 | DOMINGUEZ A A, LIM W A, QI L S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation[J]. Nature Reviews Molecular Cell Biology, 2016, 17(1): 5-15. |
| 100 | JOUNG J, KONERMANN S, GOOTENBERG J S, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening[J]. Nature Protocols, 2017, 12(4): 828-863. |
| 101 | TAK Y E, KLEINSTIVER B P, NUNEZ J K, et al. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors[J]. Nature Methods, 2017, 14(12): 1163-1166. |
| 102 | LU A R, WANG J, SUN W, et al. Reprogrammable CRISPR/dCas9-based recruitment of DNMT1 for site-specific DNA demethylation and gene regulation[J]. Cell Discovery, 2019, 5: 22. |
| 103 | LU Z H, YANG S, YUAN X, et al. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis [J]. Nucleic Acids Research, 2019, 47(7): e40. |
| 1 | POWERS-FLETCHER M V, KENDALL B A, GRIFFIN A T, et al. Filamentous fungi[J]. Microbiology Spectrum, 2016, 4(3): 1-29. |
| 2 | GILES C, LAMONT-FRIEDRICH S J, MICHL T D, et al. The importance of fungal pathogens and antifungal coatings in medical device infections[J]. Biotechnology Advances, 2018, 36(1):264-280. |
| 3 | WARD O P. Production of recombinant proteins by filamentous fungi[J]. Biotechnology Advances, 2012, 30(5): 1119-1139. |
| 4 | YANG L, LUBECK M, LUBECK P S. Aspergillus as a versatile cell factory for organic acid production[J]. Fungal Biology Reviews, 2017, 31(1):33-49. |
| 5 | KUN R S, GOMES A C S, HILDÉN K S, et al. Developments and opportunities in fungal strain engineering for the production of novel enzymes and enzyme cocktails for plant biomass degradation[J]. Biotechnology Advances, 2019, 37(6): 107361. |
| 6 | GUPTA V K, STEINDORFF A S, DE PAULA R G, et al. The post-genomic era of Trichoderma reesei: what's Next?[J]. Trends in Biotechnology, 2016, 34(12):970-982. |
| 7 | CAIRNS T C, NAI C, MEYER V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research[J]. Fungal Biology and Biotechnology, 2018, 5(1): 13. |
| 8 | HSU P D, LANDER E S, ZHANG F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6):1262-1278. |
| 9 | DOUDNA J A, CHARPENTIER E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213):1258096. |
| 10 | SHALEM O, SANJANA N E, ZHANG F. High-throughput functional genomics using CRISPR-Cas9[J]. Nature Reviews Genetics, 2015, 16(5): 299-311. |
| 11 | WANG F Y, QI L S. Applications of CRISPR genome engineering in cell biology[J]. Trends in Cell Biology, 2016, 26(11):875-888. |
| 12 | WRIGHT A V, NUNEZ J K, DOUDNA J A. Biology and applications of CRISPR Systems: harnessing nature's toolbox for genome engineering[J]. Cell, 2016, 164(1-2):29-44. |
| 13 | BARRANGOU R, DOUDNA J A. Applications of CRISPR technologies in research and beyond[J]. Nature Biotechnology, 2016, 34(9): 933-941. |
| 14 | DOUDNA J A. The promise and challenge of therapeutic genome editing[J]. Nature, 2020, 578(7794):229-236. |
| 15 | HU J H, DAVIS K M, LIU D R. Chemical biology approaches to genome editing: understanding, controlling, and delivering programmable nucleases[J]. Cell Chemical Biology, 2016, 23(1): 57-73. |
| 16 | KOMOR A C, BADRAN A H, LIU D R. CRISPR-based technologies for the manipulation of eukaryotic genomes[J]. Cell, 2017, 168(1/2): 20-36. |
| 17 | SHRIVASTAV M, DE HARO L P, NICKOLOFF J A. Regulation of DNA double-strand break repair pathway choice[J]. Cell Research, 2008, 18(1):134-147. |
| 18 | MEYER V. Genetic engineering of filamentous fungi—progress, obstacles and future trends[J]. Biotechnology Advances, 2008, 26(2):177-185. |
| 19 | PORTEUS M H, BALTIMORE D. Chimeric nucleases stimulate gene targeting in human cells[J]. Science, 2003, 300(5620):763. |
| 20 | KLUG A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation[J]. Annual Review of Biochemistry, 2010, 79:213-231. |
| 21 | MOSCOU M J, BOGDANOVE A J. A simple cipher governs DNA recognition by TAL effectors[J]. Science, 2009, 326(5959):1501. |
| 22 | CHEN K L, GAO C X. TALENs: customizable molecular DNA scissors for genome engineering of plants[J]. Journal of Genetics and Genomics, 2013, 40(6):271-279. |
| 23 | ARAZOE T, OGAWA T, MIYOSHI K, et al. Tailor-made TALEN system for highly efficient targeted gene replacement in the rice blast fungus[J]. Biotechnology and Bioengineering, 2015, 112(7): 1335-1342. |
| 24 | ISHINO Y, SHINAGAWA H, MAKINO K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J]. Journal of Bacteriology, 1987, 169(12):5429-5433. |
| 25 | BARRANGOU R, FREMAUX C, DEVEAU H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819):1709-1712. |
| 26 | KUNIN V, SOREK R, HUGENHOLTZ P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats[J]. Genome Biology, 2007, 8(4): R61. |
| 27 | LILLESTOL R K, SHAH S A, BRUGGER K, et al. CRISPR families of the crenarchaeal genus sulfolobus: bidirectional transcription and dynamic properties[J]. Molecular Microbiology, 2009, 72(1): 259-272. |
| 28 | SAPRANAUSKAS R, GASIUNAS G, FREMAUX C, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli [J]. Nucleic Acids Research, 2011, 39(21): 9275-9282. |
| 29 | DELTCHEVA E, CHYLINSKI K, SHARMA C M, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III[J]. Nature, 2011, 471(7340): 602-607. |
| 30 | DEVEAU H, GARNEAU J E, MOINEAU S. CRISPR/Cas system and its role in phage-bacteria interactions[J]. Annual Review of Microbiology, 2010, 64: 475-493. |
| 31 | GARNEAU J E, DUPUIS M È, VILLION M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA[J]. Nature, 2010, 468(7320): 67-71. |
| 32 | MOHANRAJU P, MAKAROVA K S, ZETSCHE B, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems[J]. Science, 2016, 353(6299): aad5147. |
| 33 | KOONIN E V, MAKAROVA K S, ZHANG F. Diversity, classification and evolution of CRISPR-Cas systems[J]. Current Opinion in Microbiology, 2017, 37: 67-78. |
| 34 | SHMAKOV S, SMARGON A, SCOTT D, et al. Diversity and evolution of class 2 CRISPR-Cas systems[J]. Nature Reviews Microbiology, 2017, 15(3): 169-182. |
| 35 | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| 36 | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| 37 | MALI P, YANG L, ESVELT K M, et al. RNA-guided human genome engineering via Cas9[J]. Science, 2013, 339(6121): 823-826. |
| 38 | ZETSCHE B, GOOTENBERG J S, ABUDAYYEH O O, et al. Cpf1 is a single RNA-guided endonuclease of a Class 2 CRISPR-Cas system[J]. Cell, 2015, 163(3): 759-771. |
| 39 | ZETSCHE B, HEIDENREICH M, MOHANRAJU P, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array[J]. Nature Biotechnology, 2017, 35(1): 31-34. |
| 40 | TANG X, LOWDER L G, ZHANG T, et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants[J]. Nature Plants, 2017, 3: 17018. |
| 41 | JIANG Y, QIAN F, YANG J, et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum [J]. Nature Communications, 2017, 8: 15179. |
| 42 | LEI C, LI S Y, LIU J K, et al. The CCTL (Cpf1-assisted Cutting and Taq DNA ligase-assisted Ligation) method for efficient editing of large DNA constructs in vitro [J]. Nucleic Acids Research, 2017, 45(9): e74. |
| 43 | SWIAT M A, DASHKO S, RIDDER M DEN, et al. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae [J]. Nucleic Acids Research, 2017, 45(21): 12585-12598. |
| 44 | JAVED M R, SADAF M, AHMED T, et al. CRISPR-cas system: history and prospects as a genome editing tool in microorganisms[J]. Current Microbiology, 2018, 75(12): 1675-1683. |
| 45 | DENG H, GAO R, LIAO X, et al. CRISPR system in filamentous fungi: current achievements and future directions[J]. Gene, 2017, 627(5): 212-221. |
| 46 | WANG S X, CHEN H Q, TANG X, et al. Molecular tools for gene manipulation in filamentous fungi[J]. Applied Microbiology and Biotechnology, 2017, 101(22): 8063-8075. |
| 47 | SHI T Q, LIU G N, JI R Y, et al. CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art[J]. Applied Microbiology and Biotechnology, 2017, 101(20): 7435-7443. |
| 48 | SCHUSTER M, KAHMANN R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes[J]. Fungal Genetics and Biology, 2019, 130: 43-53. |
| 49 | ZHANG S J, GUO F, YAN W, et al. Recent Advances of CRISPR/Cas9-based genetic engineering and transcriptional regulation in industrial Biology[J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 459. |
| 50 | SONG R J, ZHAI Q, SUN L, et al. CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective[J]. Applied Microbiology and Biotechnology, 2019, 103(17): 6919-6932. |
| 51 | MATSU-URA T, BAEK M, KWON J, et al. Efficient gene editing in Neurospora crassa with CRISPR technology[J]. Fungal Biology and Biotechnology, 2015, 2: 4. |
| 52 | NØDVIG C S, NIELSEN J B, KOGLE M E, et al. A CRISPR-Cas9 system for genetic engineering of filamentous fungi[J]. PLoS One, 2015, 10(7): e0133085. |
| 53 | NØDVIG C S, HOOF J B, KOGLE M E, et al. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli [J]. Fungal Genetics and Biology, 2018, 115: 78-89. |
| 54 | ZHENG X M, ZHENG P, SUN J, et al. Heterologous and endogenous U6 snRNA promoters enable CRISPR/Cas9 mediated genome editing in Aspergillus niger [J]. Fungal Biology and Biotechnology, 2018, 5: 2. |
| 55 | ZHENG X M, ZHENG P, ZHANG K, et al. 5S rRNA Promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger [J]. ACS Synthetic Biology, 2019, 8(7): 1568-1574. |
| 56 | DONG H, ZHENG J, YU D, et al. Efficient genome editing in Aspergillus niger with an improved recyclable CRISPR-HDR toolbox and its application in introducing multiple copies of heterologous genes[J]. Journal of Microbiological Methods, 2019, 163: 105655. |
| 57 | DONG L B, LIN X, YU D, et al. High-level expression of highly active and thermostable trehalase from Myceliophthora thermophila in Aspergillus niger by using the CRISPR/Cas9 tool and its application in ethanol fermentation[J]. Journal of Industrial Microbiology & Biotechnology, 2020, 47(1): 133-144. |
| 58 | HUANG L G, DONG H, ZHENG J, et al. Highly efficient single base editing in Aspergillus niger with CRISPR/Cas9 cytidine deaminase fusion[J]. Microbiological Research, 2019, 223-225: 44-50. |
| 59 | 田朝光, 李金根, 顾淑莹, 等. 一种调控sgRNA转录的启动子、表达载体,及其基因组编辑系统和应用: 201910834799.7[P]. 2019-12-10. |
| TIAN C G, LI J G, GU S Y, et al. A promoter and expression vector that regulate the transcription of sgRNA and its genome editing system and application: 201910834799.7[P]. 2019-12-10. | |
| 60 | VANEGAS K G, JARCZYNSKA Z D, STRUCKO T, et al. Cpf1 enables fast and efficient genome editing in Aspergilli [J]. Fungal Biology and Biotechnology, 2019, 6: 6. |
| 61 | KATAYAMA T, TANAKA Y, OKABE T, et al. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae [J]. Biotechnology Letters, 2016, 38(4): 637-642. |
| 62 | KATAYAMA T, NAKAMURA H, ZHANG Y, et al. Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae [J]. Applied and Environmental Microbiology, 2019, 85(3): e01896-18. |
| 63 | LIU R, CHEN L, JIANG Y, et al. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system[J]. Cell Discovery, 2015, 1: 15007. |
| 64 | HAO Z Z, SU X Y. Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex[J]. BMC Biotechnology, 2019, 19(1): 2. |
| 65 | WU C, CHEN Y, QIU Y, et al. A simple approach to mediate genome editing in the filamentous fungus Trichoderma reesei by CRISPR/Cas9-coupled in vivo gRNA transcription[J]. Biotechnology Letters, 2020, 42(7):1203-1210. |
| 66 | ZOU G, XIAO M, CHAI S, et al. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents[J]. Microbial Biotechnology, 2020, 42(7):1203-1210. |
| 67 | POHL C, KIEL J A, DRIESSEN A J, et al. CRISPR/Cas9 based genome editing of Penicillium chrysogenum [J]. ACS Synthetic Biology, 2016, 5(7): 754-764. |
| 68 | POHL C, MOZSIK L, DRIESSEN A J M, et al. Genome editing in Penicillium chrysogenum using Cas9 ribonucleoprotein particles[J]. Methods in Molecular Biology, 2018, 1772: 213-232. |
| 69 | VISSER H, JOOSTEN V, PUNT P J, et al. Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1[J]. Industial Biotechnology, 2011, 7(3): 10. |
| 70 | BERKA R M, GRIGORIEV I V, OTILLAR R, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris [J]. Nature Biotechnology, 2011, 29(10): 922-927. |
| 71 | KOLBUSZ M A, DI FALCO M, ISHMAEL N, et al. Transcriptome and exoproteome analysis of utilization of plant-derived biomass by Myceliophthora thermophila [J]. Fungal Genetics and Biology, 2014, 72: 10-20. |
| 72 | WANG J, WU Y, GONG Y, et al. Enhancing xylanase production in the thermophilic fungus Myceliophthora thermophila by homologous overexpression of Mtxyr1 [J]. Journal of Industrial Microbiology & Biotechnology, 2015, 42(9): 1233-1241. |
| 73 | YANG F, GONG Y, LIU G, et al. Enhancing cellulase production in thermophilic fungus Myceliophthora thermophila ATCC42464 by RNA interference of cre1 gene expression[J]. Journal of Microbiology and Biotechnology, 2015, 25(7): 1101-1107. |
| 74 | XU J, LI J, LIN L, et al. Development of genetic tools for Myceliophthora thermophila [J]. BMC Biotechnology, 2015, 15: 35. |
| 75 | LIU Q, GAO R, LI J, et al. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering[J]. Biotechnology for Biofuels, 2017, 10: 1. |
| 76 | KWON M J, SCHUTZE T, SPOHNER S, et al. Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi[J]. Fungal Biology and Biotechnology, 2019, 6: 15. |
| 77 | LIU Q, ZHANG Y, LI F, et al. Upgrading of efficient and scalable CRISPR-Cas-mediated technology for genetic engineering in thermophilic fungus Myceliophthora thermophila [J]. Biotechnology for Biofuels, 2019, 12: 293. |
| 78 | JIMENEZ A, MUNOZ-FERNANDEZ G, LEDESMA-AMARO R, et al. One-vector CRISPR/Cas9 genome engineering of the industrial fungus Ashbya gossypii [J]. Microbial Biotechnology, 2019, 12(6): 1293-1301. |
| 79 | JIMENEZ A, HOFF B, REVUELTA J L. Multiplex genome editing in Ashbya gossypii using CRISPR-Cpf1[J]. New Biotechnology, 2020, 57: 29-33. |
| 80 | ARAZOE T, MIYOSHI K, YAMATO T, et al. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus[J]. Biotechnology and Bioengineering, 2015, 112(12): 2543-2549. |
| 81 | FOSTER A J, MARTIN-URDIROZ M, YAN X, et al. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus[J]. Scientific Reports, 2018, 8(1): 14355. |
| 82 | YAMATO T, HANDA A, ARAZOE T, et al. Single crossover-mediated targeted nucleotide substitution and knock-in strategies with CRISPR/Cas9 system in the rice blast fungus[J]. Scientific Reports, 2019, 9(1): 7427. |
| 83 | FULLER K K, CHEN S, LOROS J J, et al. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus [J]. Eukaryotic Cell, 2015, 14(11): 1073-1080. |
| 84 | ZHANG C, MENG X, WEI X, et al. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus [J]. Fungal Genetics and Biology, 2016, 86: 47-57. |
| 85 | WEBER J, VALIANTE V, NØDVIG C S, et al. Functional reconstitution of a fungal natural product gene cluster by advanced genome editing[J]. ACS Synthetic Biology, 2017, 6(1): 62-68. |
| 86 | SCHUSTER M, SCHWEIZER G, REISSMANN S, et al. Genome editing in Ustilago maydis using the CRISPR-Cas system[J]. Fungal Genetics and Biology, 2016, 89: 3-9. |
| 87 | CHEN J J, LAI Y L, WANG L L, et al. CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus Beauveria bassiana [J]. Scientific Reports, 2017, 7(1): 45763. |
| [1] | 董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117. |
| [2] | 陈盈盈, 刘扬, 史俊杰, 马俊英, 鞠建华. CRISPR/Cas基因编辑及其新兴技术在丝状真菌研究中的系统应用[J]. 合成生物学, 2024, 5(3): 672-693. |
| [3] | 杜瑶, 高宏丹, 刘家坤, 刘孝荣, 邢志浩, 张涛, 马东礼. CRISPR-Cas系统在病原核酸检测中的研究进展[J]. 合成生物学, 2024, 5(1): 202-216. |
| [4] | 林继聪, 邹根, 刘宏民, 魏勇军. CRISPR/Cas基因组编辑技术在丝状真菌次级代谢产物合成中的应用[J]. 合成生物学, 2023, 4(4): 738-755. |
| [5] | 马孟丹, 刘宇辰. 合成生物学在疾病信息记录与实时监测中的应用潜力[J]. 合成生物学, 2023, 4(2): 301-317. |
| [6] | 肖晗, 刘宜欣. CRISPR-Cas系统编辑丝状真菌的进展与挑战[J]. 合成生物学, 2021, 2(2): 274-286. |
| [7] | 黄雪年, 唐慎, 吕雪峰. 工业丝状真菌土曲霉合成生物技术研究进展及展望[J]. 合成生物学, 2020, 1(2): 187-211. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||