合成生物学 ›› 2021, Vol. 2 ›› Issue (3): 323-334.DOI: 10.12211/2096-8280.2020-086
DNA数字信息存储的研究进展
董一名1, 孙法家1, 武瑞君2, 钱珑1
- 1.北京大学定量生物学中心,北京 100871
2.中国生物技术发展中心战略与政策处,北京 100039
-
收稿日期:2020-11-30修回日期:2021-04-04出版日期:2021-06-30发布日期:2021-07-13 -
通讯作者:钱珑 -
作者简介:董一名 (1993—),女,博士研究生。研究方向为合成生物学。E-mail:ymdong@pku.edu.cn孙法家 (1996—),男,博士研究生。研究方向为合成生物学。E-mail:projectyasuo@pku.edu.cn钱珑 (1985—),女,助理研究员。研究方向为进化系统生物学、生物信息学和合成生物学。E-mail:long.qian@pku.edu.cn -
基金资助:国家自然科学基金(31901063);国家重点研发计划“合成生物学”重点专项(2020YFA0906900)
Research progress on DNA molecules for digital information storage
DONG Yiming1, SUN Fajia1, WU Ruijun2, QIAN Long1
- 1.Center for Quantitative Biology,Peking University,Beijing 100871,China
2.Division of Strategy and Policy,China National Center for Biotechnology Development,Beijing 100039,China
-
Received:2020-11-30Revised:2021-04-04Online:2021-06-30Published:2021-07-13 -
Contact:QIAN Long
摘要:
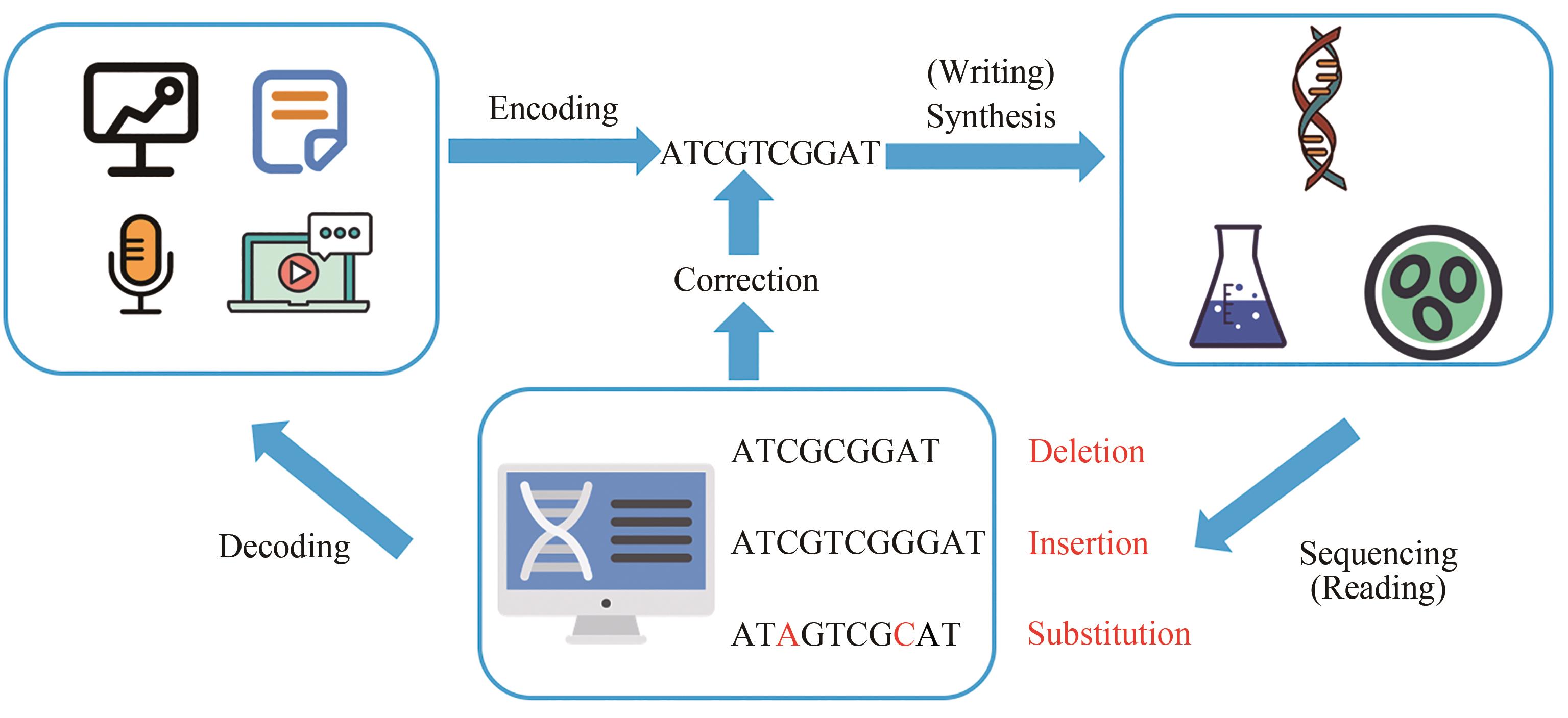
随着计算机技术的发展,数字化信息存储改变了我们的生活。信息正在以越来越快的速度产生着,但与此伴生的,是如何有效存储数据的问题。诸如磁盘、硬盘、闪存等磁学或光学等传统存储介质已经逐渐不能满足全世界范围内数据存储的需要。DNA分子凭借其稳定性、高存储密度和低维护成本,有望成为实用的新型信息存储介质。本文首先介绍了利用DNA分子进行数据存储的工作流程,继而介绍了DNA数据存储领域的研究历史和研究进展,包括存储方式、读取方式、编码方式等。为实现DNA信息存储,通过信息编码将二进制信息转换成DNA序列信息;DNA合成实现信息写入;最后通过基因测序获取序列信息,进而进行信息解码得到原始信息。而现代分子生物学技术的发展,尤其是DNA合成和测序技术的飞跃,使DNA分子大规模存储人工数据逐渐成为现实。之后,对比了DNA分子相对于传统数据存储介质的优劣,介绍了基于DNA分子的数据存储的风险与挑战,如数据安全性、信息读写的速度和成本等。最后,对DNA数据存储领域未来研究的方向进行了展望,介绍了一些与该领域具备交叉潜力的新兴生物技术领域,如“DNA条形码”“DNA折纸”。
中图分类号:
引用本文
董一名, 孙法家, 武瑞君, 钱珑. DNA数字信息存储的研究进展[J]. 合成生物学, 2021, 2(3): 323-334.
DONG Yiming, SUN Fajia, WU Ruijun, QIAN Long. Research progress on DNA molecules for digital information storage[J]. Synthetic Biology Journal, 2021, 2(3): 323-334.
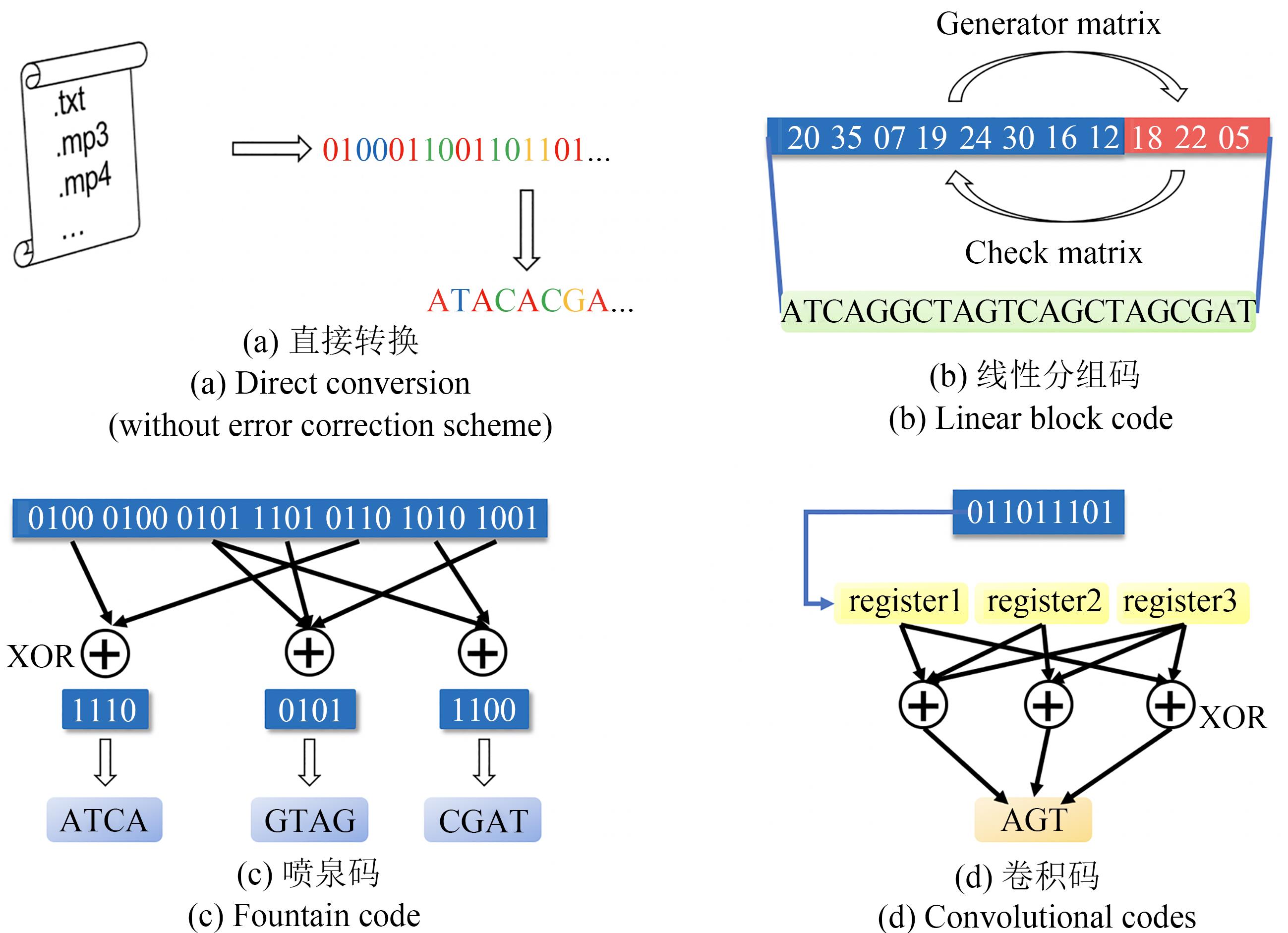
图2 DNA存储研究中使用的信息编码方法(前向纠错体系)[(a) 直接转换,不包含纠错方案。在这种方案中,数据被读取为数字流,然后转换为DNA序列。例如,Church等[28]和Goldman等[29]分别将二进制数字流和三进制数字流中的每一位转换为一个DNA碱基。(b)线性分组码,即通过线性运算,从原始信息(信息码元)产生用于纠错的冗余(称为“校验码元”或“监督码元”)。在解码时,与生成矩阵相对应的校验矩阵可以用于校验接收到的信息中是否包含错误,并进行纠正。(c) 喷泉码,即将原始信息转换为大量较短的信息,这些较短的信息并非原始信息的一部分,而是将原始信息中的符号通过特定的分布进行异或运算得到的。在解码时,只要获得了足够数量的短信息,就可以恢复原始信息。(d) 卷积码,即“有记忆”的编码方案。在编码用于传输的符号时,不仅需要处理当前的信息符号,还要对当前位置之前的数个信息符号进行运算]
Fig. 2 Information encoding method (forward error correction system) used in DNA storage research[(a) Direct conversion without error correction scheme. In this method, the data is read as a digital stream and then converted into DNA sequences. For example, Church et al.[28] and Goldman et al.[29] converted each bit in a binary number stream and a ternary number stream into a DNA base, respectively.(b) Linear block code, namely, generating redundancy for error correction (called "check symbols" or "supervision symbols") from the original information (information symbols) through linear operations. In the decoding process, the check matrix corresponding to the generator matrix can be used to check whether the received information contains errors and then correct them. (c) Fountain code, which converts the original information into a large number of shorter sequences. These shorter sequences are not part of the original information, but obtained by performing XOR operations on the symbols in the original information according to a specific distribution. In the decoding process, as long as a sufficient number of shorter sequences are obtained, the original information can be restored. (d) Convolutional codes, that is, coding schemes "with memory". Both the current information symbol and several information symbols before the current position are used to generate the encoding symbols]
| 1 | BOHANNON J. DNA: the ultimate hard drive[EB/OL]. [2012-08-16]. . |
| 2 | The Economic Times. Global data to increase 10x by 2025: data age 2025[EB/OL]. [2017-04-04]. . |
| 3 | World Semiconductor Trade Statistics. WSTS semiconductor market forecast autumn 2020 [EB/OL].[2020-12-01]. . |
| 4 | WATSON J D, CRICK F H. Molecular structure of nucleic acids:a structure for deoxyribose nucleic acid[J]. Nature, 1953, 248(4): 623-624. |
| 5 | CRICK F. Central dogma of molecular biology[J]. Nature, 1970, 227: 561-563. |
| 6 | SHRIVASTAVA S, BADLANI R. Data storage in DNA[J]. International Journal of Electrical Power & Energy Systems, 2014, 2: 119-124. |
| 7 | EXTANCE A. How DNA could store all the world's data[J]. Nature, 2016, 537: 22-24. |
| 8 | ALLENTOFT M E, COLLINS M, HARKER D, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils[J]. Proceedings Biological Sciences, 2012, 279(1748): 4724-4733. |
| 9 | RUTTEN M G T A, VAANDRAGER F W, ELEMANS J A A W, et al. Encoding information into polymers[J]. Nature Reviews Chemistry, 2018, 2: 365-381. |
| 10 | PING Z, MA D Z, HUANG X L, et al. Carbon-based archiving: current progress and future prospects of DNA-based data storage[J]. GigaScience, 2019, 8(6): giz075. |
| 11 | DONG Y M, SUN F J, PING Z, et al. DNA storage: research landscape and future prospects[J]. National Science Review, 2020, 7(6): 1092-1107. |
| 12 | SEKERKA R F. Entropy and information theory[J]. Thermal Physics, 2015, 11: 247-256. |
| 13 | SHANNON C E. Prediction and entropy of printed English[J]. The Bell System Technology Journal, 1951, 30(1): 50-64. |
| 14 | YAZDI S M, YUAN Y, MA J, et al. A rewritable, random-access DNA-based storage system[J]. Scientific Reports, 2015, 5: 14138. |
| 15 | MIZUOCHI T. Recent progress in forward error correction and its interplay with transmission impairments[J]. IEEE Journal of Selected Topics in Quantum Electronics, 2006, 12(4): 544-554. |
| 16 | NAFAA A, TALEB T, MURPHY L. Forward error correction strategies for media streaming over wireless networks[J]. IEEE Communications Magazine, 2008, 46(1): 72-79. |
| 17 | HAMMING R W. Error detecting and error correcting codes[J]. The Bell System Technical Journal, 1950, 23(2): 147-160. |
| 18 | BOSE R C, RAY-CHAUDHURI D K. On a class of error correcting binary group codes[J]. Information and Control, 1960, 3(1): 68-79. |
| 19 | HOCQUENGHEM A. Codes correcteurs d' erreurs[J]. Chiffres, 1959, 2: 147-156. |
| 20 | REED I S, SOLOMON G. Polynomial codes over certain finite fields[J]. Journal of the Society for Industrial and Applied Mathematics, 1960, 8(2): 300-304. |
| 21 | BYERS J W, LUBY M, MITZENMACHER M. A digital fountain approach to reliable distribution of bulk data[C]// Proceedings of the ACM SIGCOMM' 98 Conference on Applications, Technologies, Architectures, and Protocols for Computer Communication. California:Systems Research Center, 1998, 28(4): 56-67. |
| 22 | LUBY M. LT code[C]// Proceeding of the 43rd Annual IEEE Symposium on Foundations of Computer Science. Vancouver:TCMF, 2002: 271-282. |
| 23 | HUTCHINSON R, ROSENTHAL J, SMARANDACHE R. Convolutional codes with maximum distance profile[J]. Systems & Control Letters, 2003, 54(1): 53-63. |
| 24 | ALMEIDA P, NAPP D, PINTO R. A new class of superregular matrices and MDP convolutional codes[J]. Linear Algebra and its Applications, 2013, 439(7): 2145-2157. |
| 25 | PUCHINGER S, RENNER J, ROSENKILDE J. Generic decoding in the sum-rank metric[C]//2020 IEEE International Symposium on Information Theory (ISIT). Los Angeles: Institute of Electrical and Electronics Engineering, 2020:54-59. |
| 26 | NAPP D, PINTO R, ROSENTHAL J, et al.MRD rank metric convolutional codes[C]//2017 IEEE International Symposium on Information Theory (ISIT). Aachen: Institute of Electrical and Electronics Engineering, 2017: 2766-2770. |
| 27 | ALMEIDA P, NAPP D, PINTO R. Superregular matrices and applications to convolutional codes[J]. Linear Algebra and Its Applications, 2016, 499: 1-25. |
| 28 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA[J]. Science, 2012, 337: 1628. |
| 29 | GOLDMAN N, BERTONE P, CHEN S, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA[J]. Nature, 2013, 494: 77-79. |
| 30 | LEPROUST E M, PECK B J, SPIRIN K, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process[J]. Nucleic Acids Research, 2010, 38(8): 2522-2540. |
| 31 | CARUTHERS M H. The chemical synthesis of DNA/RNA: our gift to science[J]. The Journal of Biological Chemistry, 2013, 288(2): 1420-1427. |
| 32 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications[J]. Nature Methods, 2014, 11(5): 499-507. |
| 33 | LEE H H, KALHOR R, GOELA N, et al. Terminator-free template-independent enzymatic DNA synthesis for digital information storage[J]. Nature Communications, 2019, 10(1): 2383. |
| 34 | SANGER F, NICKLEN S, COULSON A R. DNA sequencing with chain-terminating inhibitors[J]. Proceedings of the National Academy of Sciences of the United States of America, 1977, 74(12): 5463-5467. |
| 35 | WETTERSTRAND K A. DNA sequencing costs: data from the NHGRI Genome Sequencing Program (GSP)[EB/OL]. National Human Genome Research Institute.[2021-05-11]. . |
| 36 | DAHM R. Discovering DNA: Friedrich Miescher and the early years of nucleic acid research[J]. Human Genetics, 2008, 122(6): 565-581. |
| 37 | KOSSEL A. Ueber das Nucleïn der Hefe[J]. Zeitschrift für physiologische Chemie, 1879, 3(4): 284-291. |
| 38 | AVERY O T, MACLEOD C M, MCCARTY M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type iii[J]. The Journal of Experimental Medicine, 1944, 79(2): 137-158. |
| 39 | HERSHEY A D, CHASE M. Independent functions of viral protein and nucleic acid in growth of bacteriophage[J]. Journal of General Physiology, 1952, 36(1): 39-56. |
| 40 | DAVIS J. Microvenus[J]. Art Journal, 1996, 55: 70-74. |
| 41 | BANCROFT C, BOWLER T, BLOOM B, et al. Long-term storage of information in DNA[J]. Science, 2001, 293: 1763-1765. |
| 42 | GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes[J]. Angewandte Chemie International Edition, 2015, 54(8): 2552-2555. |
| 43 | BLAWAT M, GAEDKE K, HUETTER I, et al. Forward error correction for DNA data storage[J]. Procedia Computer Science, 2016, 80: 1011-1022. |
| 44 | BORNHOLT J, LOPEZ R, CARMEAN D M, et al. A DNA-based archival storage system[J]. IEEE Micro, 2017, 99: 1. |
| 45 | ERLICH Y, ZIELINSKI D, ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture[J]. Science, 2017, 355(6328): 950-954. |
| 46 | SHIPMAN S L, NIVALA J, MACKLIS J D, et al. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria[J]. Nature, 2017, 547(7663): 345-349. |
| 47 | ORGANICK L, ANG S D, CHEN Y, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36: 242-248. |
| 48 | KOCH J, GANTENBEIN S, MASANIA K, et al. A DNA-of-things storage architecture to create materials with embedded memory[J]. Nature Biotechnology, 2020, 38(1): 39-43. |
| 49 | BIOGLIO V, GRANTOTO M, GAETA R, et al. On the fly Gaussian elimination for the LT Codes[J]. IEEE Communications Letters, 2009, 13(12): 953-955. |
| 50 | HAYAZNEH K F, OUSEFIS, VALIPOUR M. Improved finite-length Luby transform codes in the binary erasure channel[J]. IET Communications, 2015, 9(8): 1122-1130. |
| 51 | PRESS W H, HAWKINS J A, JONES S K, et al. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints[J]. Proceedings of the National Academy of Science of the United States of America, 2020, 117(31): 18489-18496. |
| 52 | MOTT N. Microsoft demonstrates automated DNA storage[EB/OL].[2021-05-11]. ,38902.html. |
| 53 | BENNER S A, BATTERSBY T R, ESCHGFALLER B, et al. Redesigning nucleic acids[J]. Pure and Applied Chemistry, 1998, 70(2): 263-266. |
| 54 | GEORGIADIS M M, SINGH I, KELLETT W F, et al. Structural basis for a six nucleotide genetic alphabet[J]. Journal of the American Chemical Society, 2015, 137(21): 6947-6955. |
| 55 | ZHANG L Q, YANG Z Y, SEFAH K, et al. Evolution of functional six-nucleotide DNA[J]. Journal of the American Chemical Society, 2015, 137(21): 6734-6737. |
| 56 | HOSHIKA S, LEAL N A, KIM M J, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks[J]. Science, 2019, 363: 884-887. |
| 57 | ANAVY L, VAKNIN I, ATAR O, et al. Data storage in DNA with fewer synthesis cycles using composite DNA letters[J]. Nature Biotechnology, 2019, 37(10): 1229-1236. |
| 58 | CHOI Y, RYU T, LEE A C, et al. High information capacity DNA-based data storage with augmented encoding characters using degenerate bases[J]. Scientific Reports, 2019, 9(1): 6582. |
| 59 | LEE W, ZHOU Z, CHEN X, et al. A rewritable optical storage medium of silk proteins using near-field nano-optics[J]. Nature Nanotechnology, 2020, 15: 941-947. |
| 60 | KENNEDY E, ARCADIA C E, GEISER J, et al. Encoding information in synthetic metabolomes[J]. PLoS One, 2019, 14(7): e0217364. |
| 61 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 62 | HAJIBABAEI M, SINGER G A, HEBERT P D, et al. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics[J]. Trends in Genetics, 2007, 23(4): 167-172. |
| 63 | QIAN J, LU Z X, MANCUSO C P, et al. Barcoded microbial system for high-resolution object provenance[J]. Science, 2020, 368(6495): 1135-1140. |
| 64 | ROGERS Z N, MCFARLAND C D, WINTERS I P, et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice[J]. Nature Genetics, 2018, 50(4): 483-486. |
| 65 | WIRTH D, GAMA-NORTON L, RIEMER P, et al. Road to precision: recombinase-based targeting technologies for genome engineering[J]. Current Opinion in Biotechnology, 2007, 18(5): 411-419. |
| 66 | GRINDLEY N D F, WHITESON K L, RICE P A. Mechanisms of site-specific recombination[J]. Annual Review of Biochemistry, 2006, 75: 567-605. |
| 67 | KIM J, BAE J H, BAYM M, et al. Metastable hybridization-based DNA information storage to allow rapid and permanent erasure[J]. Nature Communications, 2020, 11(1): 5008. |
| 68 | GRASS R N, HECKEL R, DESSIMOZ C, et al. Genomic encryption of digital data stored in synthetic DNA[J]. Angewandte Chemie International Edition, 2020, 59(22): 8476-8480. |
| 69 | ZHANG Y, MAO X, LI F, et al. Nanoparticle-assisted alignment of carbon nanotubes on DNA origami[J]. Angewandte Chemie International Edition, 2020, 59(12): 4892-4896. |
| 70 | LIU X, ZHANG F, JING X, et al. Complex silica composite nanomaterials templated with DNA origami[J]. Nature, 2018, 559(7715): 593-598. |
| 71 | LOMAN N J, QUICK J, SIMPSON J T. A complete bacterial genome assembled de novo using only nanopore sequencing data[J]. Nature Methods, 2015, 12(8): 733-735. |
| 72 | JAIN M, FIDDES I T, MIGA K H, et al. Improved data analysis for the MinION nanopore sequencer[J]. Nature Methods, 2015, 12(4): 351-356. |
| 73 | LAVER T, HARRISON J, O'NEILL P A, et al. Assessing the performance of the Oxford nanopore technologies MinION[J]. Biomolecular Detection and Quantification, 2015, 3: 1-8. |
| 74 | QUAIL M A, SMITH M, COUPLAND P, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers[J]. BMC Genomics, 2012, 13(1): 341. |
| 75 | GOODWIN S, MCPHERSON J D, MCCOMBIE W R. Coming of age: ten years of next-generation sequencing technologies[J]. Nature Reviews Genetics, 2016, 17(6): 333-351. |
| 76 | ESCALONA M, ROCHA S, POSADA D. A comparison of tools for the simulation of genomic next-generation sequencing data[J]. Nature Reviews Genetics, 2016, 17: 459-469. |
| 77 | GAWAD C, KOH W, QUAKE S R. Single-cell genome sequencing: current state of the science[J]. Nature Reviews Genetics, 2016, 17: 175-188. |
| 78 | MARDIS E R. A decade's perspective on DNA sequencing technology[J]. Nature, 2011, 470(7333): 198-203. |
| 79 | LOPEZ R, CHEN Y J, DUMAS ANG S, et al. DNA assembly for nanopore data storage readout[J]. Nature Communications, 2019, 10(1): 2933. |
| 80 | FARZADFARD F, LU T K. Emerging applications for DNA writers and molecular recorders[J]. Science, 2018, 361(6405): 870-875. |
| 81 | LOMEDICO P T. Use of recombinant DNA technology to program eukaryotic cells to synthesize rat proinsulin: a rapid expression assay for cloned genes[J]. Proceedings of the National Academy of Sciences of the United States of America, 1982, 79(19): 5798-5802. |
| 82 | FARZADFARD F, LU T K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations[J]. Science, 2014, 346(6211):1256272. |
| [1] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [2] | 张宣梁, 李青婷, 王飞. DNA存储系统中的数据写入[J]. 合成生物学, 2024, 5(5): 1125-1141. |
| [3] | 陈大明, 张学博, 刘晓, 马悦, 熊燕. 从全球专利分析看DNA合成与信息存储技术发展趋势[J]. 合成生物学, 2021, 2(3): 399-411. |
| [4] | 黄小罗, 戴俊彪. 人工DNA合成技术:DNA数据存储的基石[J]. 合成生物学, 2021, 2(3): 335-353. |
| [5] | 闫汉, 肖鹏峰, 刘全俊, 陆祖宏. DNA微阵列原位化学合成[J]. 合成生物学, 2021, 2(3): 354-370. |
| [6] | 韩明哲, 陈为刚, 宋理富, 李炳志, 元英进. DNA信息存储:生命系统与信息系统的桥梁[J]. 合成生物学, 2021, 2(3): 309-322. |
| [7] | 郜艳敏, 唐梦童, 刘倩, 乔宏艳, 王桃雪, 齐浩. DNA信息存储中关键生化方法的研究[J]. 合成生物学, 2021, 2(3): 384-398. |
| [8] | 彭凯, 逯晓云, 程健, 刘莹, 江会锋, 郭晓贤. DNA合成、组装与纠错技术研究进展[J]. 合成生物学, 2020, 1(6): 697-708. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||