合成生物学 ›› 2020, Vol. 1 ›› Issue (6): 697-708.DOI: 10.12211/2096-8280.2020-034
DNA合成、组装与纠错技术研究进展
彭凯1,2, 逯晓云1, 程健1, 刘莹1, 江会锋1, 郭晓贤1
- 1.中国科学院天津工业生物技术研究所系统微生物工程重点实验室,天津 300308
2.天津科技大学生物工程学院,天津 300457
-
收稿日期:2020-03-23修回日期:2020-10-22出版日期:2020-12-31发布日期:2021-01-15 -
通讯作者:江会锋,郭晓贤 -
作者简介:彭凯(1995—),男,硕士研究生。主要研究方向为DNA纠错。E-mail: pengk@tib.cas.cn
江会锋(1981—),男,博士,研究员。主要研究方向为代谢合成生物学。E-mail: jiang_hf@tib.cas.cn
郭晓贤(1982—),男,博士,副研究员。主要研究方向为酶法DNA合成。E-mail: guoxx@tib.cas.cn -
基金资助:国家重点研发计划(2020YFC1316400);天津市合成生物技术创新能力提升行动(TSBICIP-KJGG-007-01)
Advances in technologies for de novo DNA synthesis, assembly and error correction
PENG Kai1,2, LU Xiaoyun1, CHENG Jian1, LIU Ying1, JIANG Huifeng1, GUO Xiaoxian1
- 1.Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,Tianjin 300308,China
2.School of Biological Engineering,Tianjin University of Science and Technology,Tianjin 300457,China
-
Received:2020-03-23Revised:2020-10-22Online:2020-12-31Published:2021-01-15 -
Contact:JIANG Huifeng, GUO Xiaoxian
摘要:
DNA设计合成是推动生命科学及其相关领域发展的关键共性底层技术。常规的遗传操作技术仅能对已有的DNA序列进行有限的改造,而DNA合成技术则可从头“书写”生命信息,从另一高度提升我们对生命体理解、预测和操控的能力。DNA合成技术包括寡核苷酸合成技术、DNA组装技术以及DNA纠错技术。本文总结了以上关键技术的特点和发展趋势,经历超过60年的发展后,化学合成法仍然是当前寡核苷酸合成的主流方法,它被广泛应用于柱式及芯片DNA合成仪,酶法DNA合成技术则有望颠覆传统的DNA化学合成方法;现有DNA合成技术在合成能力和准确性上存在局限,难以直接准确合成基因长度的DNA片段,分级的体外与体内组装技术的合理搭配,可将分段合成的寡核苷酸片段装配成长片段DNA,达到基因长度甚至基因组长度DNA序列的合成,它也因此成为长片段DNA合成的关键;寡核苷酸的合成与组装过程都不可避免地引入错误,基于错配结合或错配切除的纠错技术在DNA合成过程不同阶段的应用,不仅能提高DNA合成的准确性,还可有效降低长片段DNA合成的质控成本。近年来合成生物学等相关领域的迅猛发展,对DNA合成相关技术提出了新的要求,正推动DNA合成、组装与纠错相关技术向着高通量、自动化和集成化的方向不断改进和创新。
中图分类号:
引用本文
彭凯, 逯晓云, 程健, 刘莹, 江会锋, 郭晓贤. DNA合成、组装与纠错技术研究进展[J]. 合成生物学, 2020, 1(6): 697-708.
PENG Kai, LU Xiaoyun, CHENG Jian, LIU Ying, JIANG Huifeng, GUO Xiaoxian. Advances in technologies for de novo DNA synthesis, assembly and error correction[J]. Synthetic Biology Journal, 2020, 1(6): 697-708.
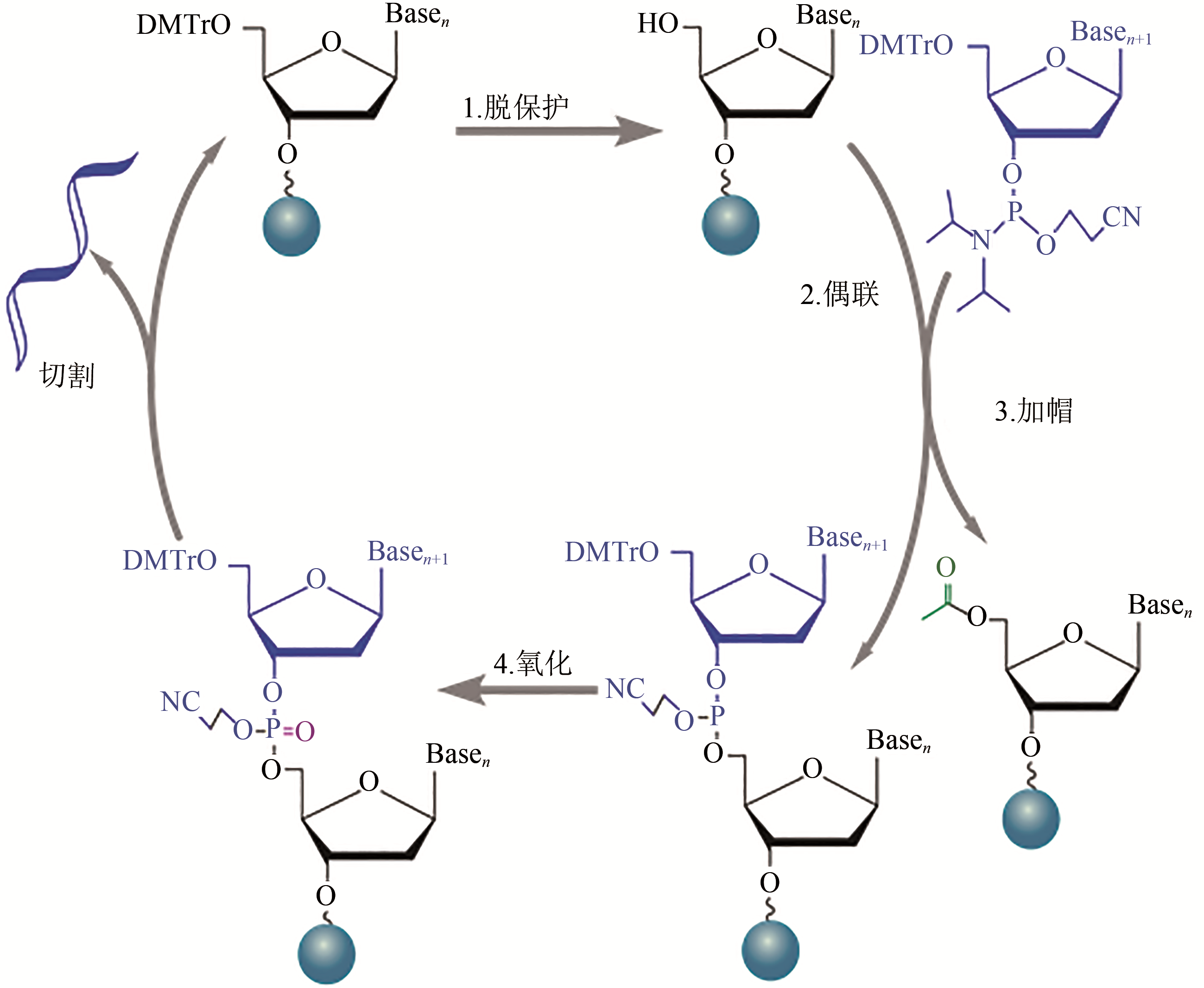
图1 固相亚磷酰胺寡核苷酸合成反应循环[1—脱保护,用三氯乙酸脱去固相载体上核苷酸5' 位的DMT(dimethoxytrityl)保护基团,获得供下一步缩合反应的游离5' 羟基;2—偶联,将亚磷酰胺保护的核苷酸单体与四氮唑活化剂混合,形成亚磷酰胺四唑活性中间体(其3' 端已被活化,但5' 端仍受DMT保护),与脱保护后生成的5' 羟基发生缩合反应,寡核苷酸链向前延长一个核苷酸;3—加帽,缩合反应后,通过乙酰化来封闭未参与反应的5' 羟基,防止其在随后的循环反应中被延伸,以减少产物中单核苷酸缺失片段的比例;4—氧化,用碘的四氢呋喃溶液将连接3',5' 的不稳定亚磷酰胺三酯氧化为磷酸三酯,得到稳定的寡核苷酸链]
Fig. 1 Reaction cycle for solid-phase phosphoramidite synthesis of oligonucleotide[1—Deprotection. The dimethoxytrityl (DMT) ether capping group on the 5’ end of the previous nucleotide is removed to generate the 5’ end hydroxyl group on the growing chain. 2—Coupling. The desired phosphoramidite monomer couples to the chain via the 5’ end hydroxyl group generated in the deprotection step. 3—Capping. Uncoupled 5’ end hydroxyl group is blocked by acylating molecules to prevent further reactions. 4—Oxidation. An oxidizer agent is used to converte phosphite triester linkage to a more stable phosphotriester bond prior to the start of the next cycle. Cleavage: final oligonucleotides are cleaved from their solid support at the end of synthesis]
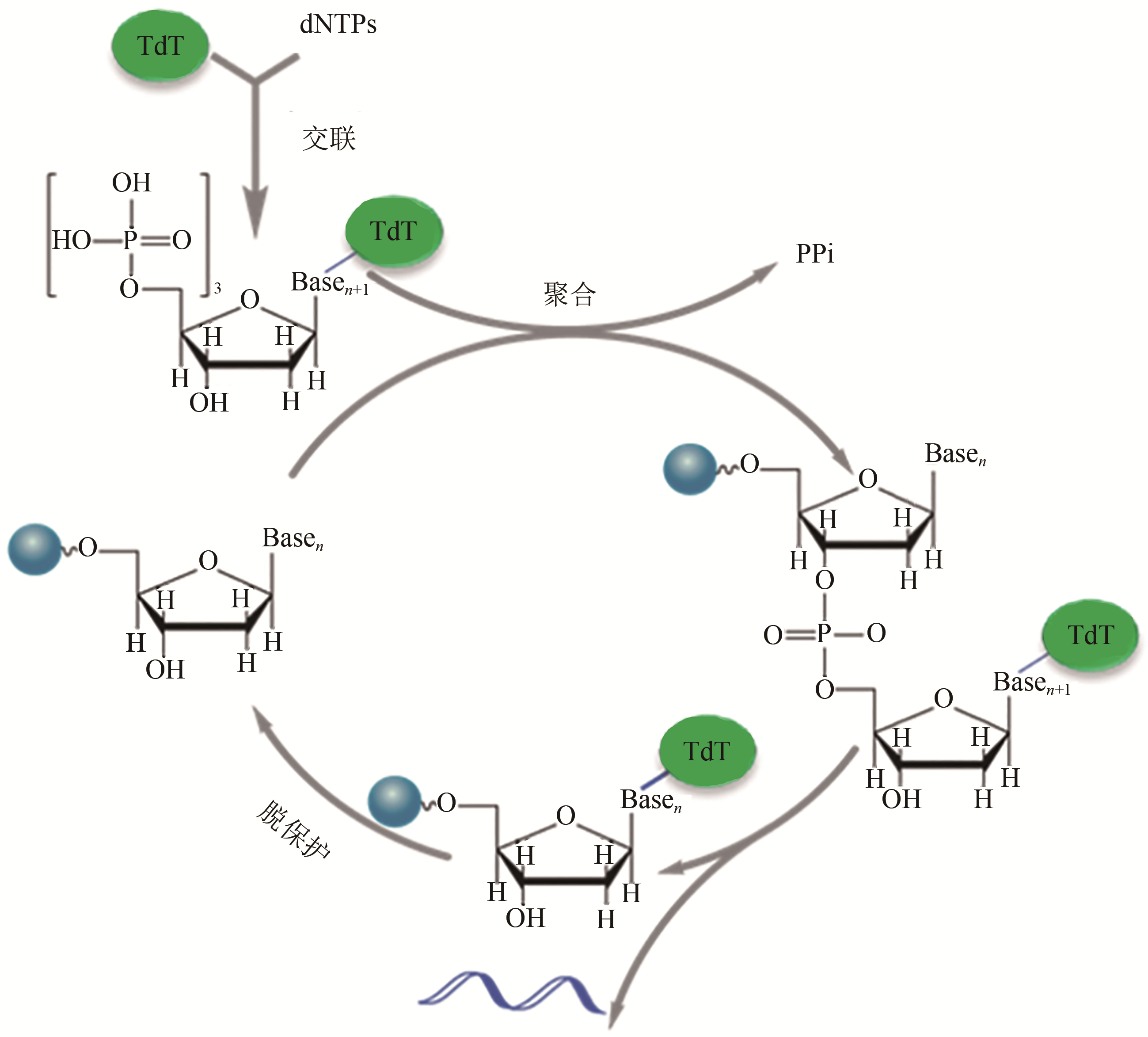
图2 TdT-dNTP 交联体介导的可逆终止用于寡核苷酸合成循环[TdT (terminal deoxynucleotidyl transferase) is specifically conjugated with the desired dNTP to form TdT-dNTP conjugate before the reaction cycle. The conjugate is then incorporated into the 3' end of the primer in the polymerization reaction. The subsequent deprotection reaction removes TdT and unreacted TdT-dNTP conjugate to release the primer for next round of polymerization. The cycle is iterated to extend a primer by a defined sequence]
Fig. 2 The oligo synthesis cycle by TdT-dNTP conjugates mediated reversible termination
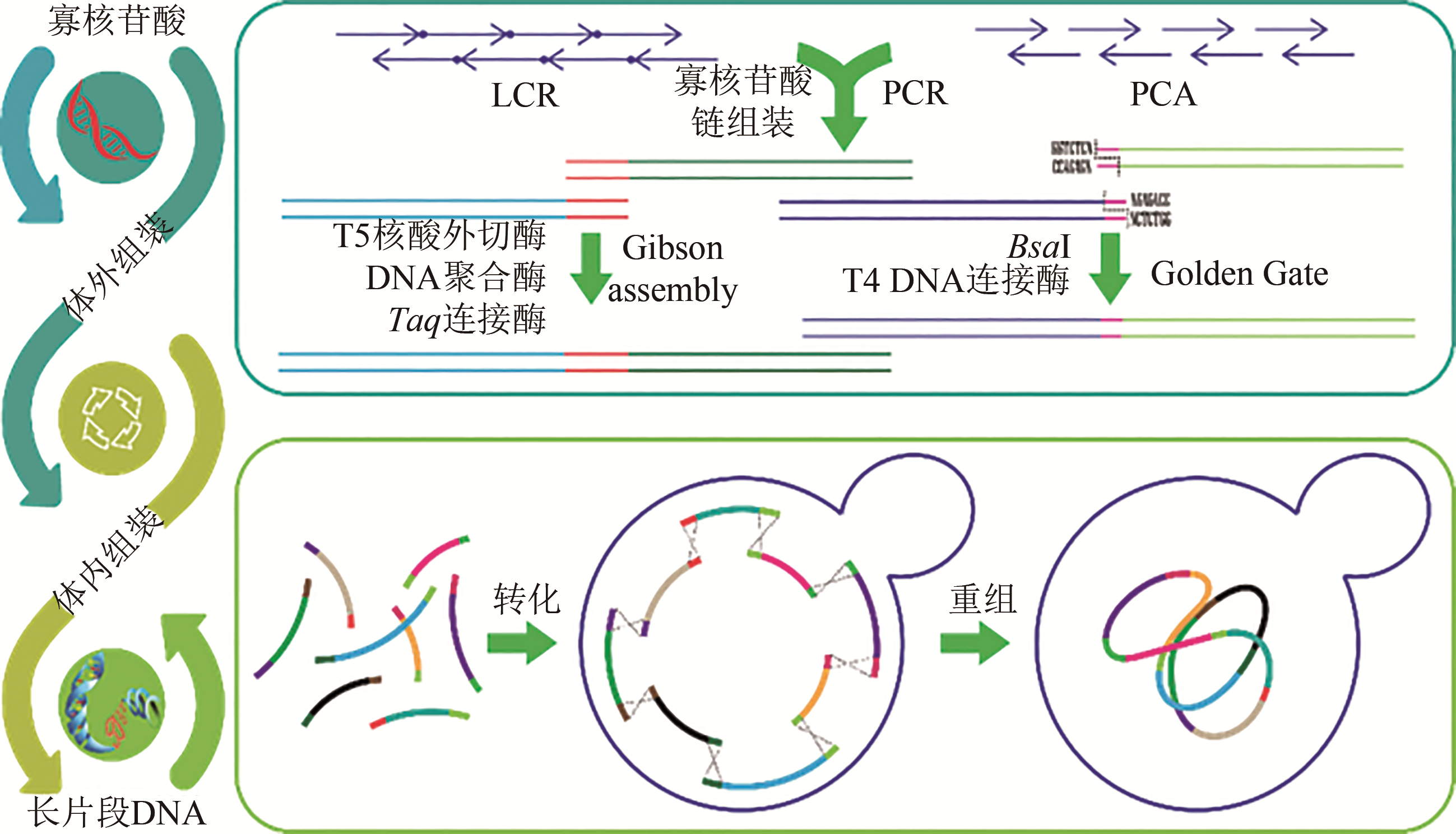
图3 常见体外和体内DNA组装技术及流程(Assembly of synthesized overlapping single-stranded oligos usually employ LCR and PCA based approaches. These assembly process turn oligos into longer double-stranded DNA constructs which can then be further assembled into longer fragments by Gibson assembly and Golden gate. In vivo assembly is typically achieved by feeding yeast cell with large fragments that have overlapping ends through transformation. The efficient DNA recombination machinery eventually string the fragments for assemblies that span hundreds of thousands or even millions of bases)
Fig. 3 Summary of general schemes of in vitro and in vivo DNA assembly
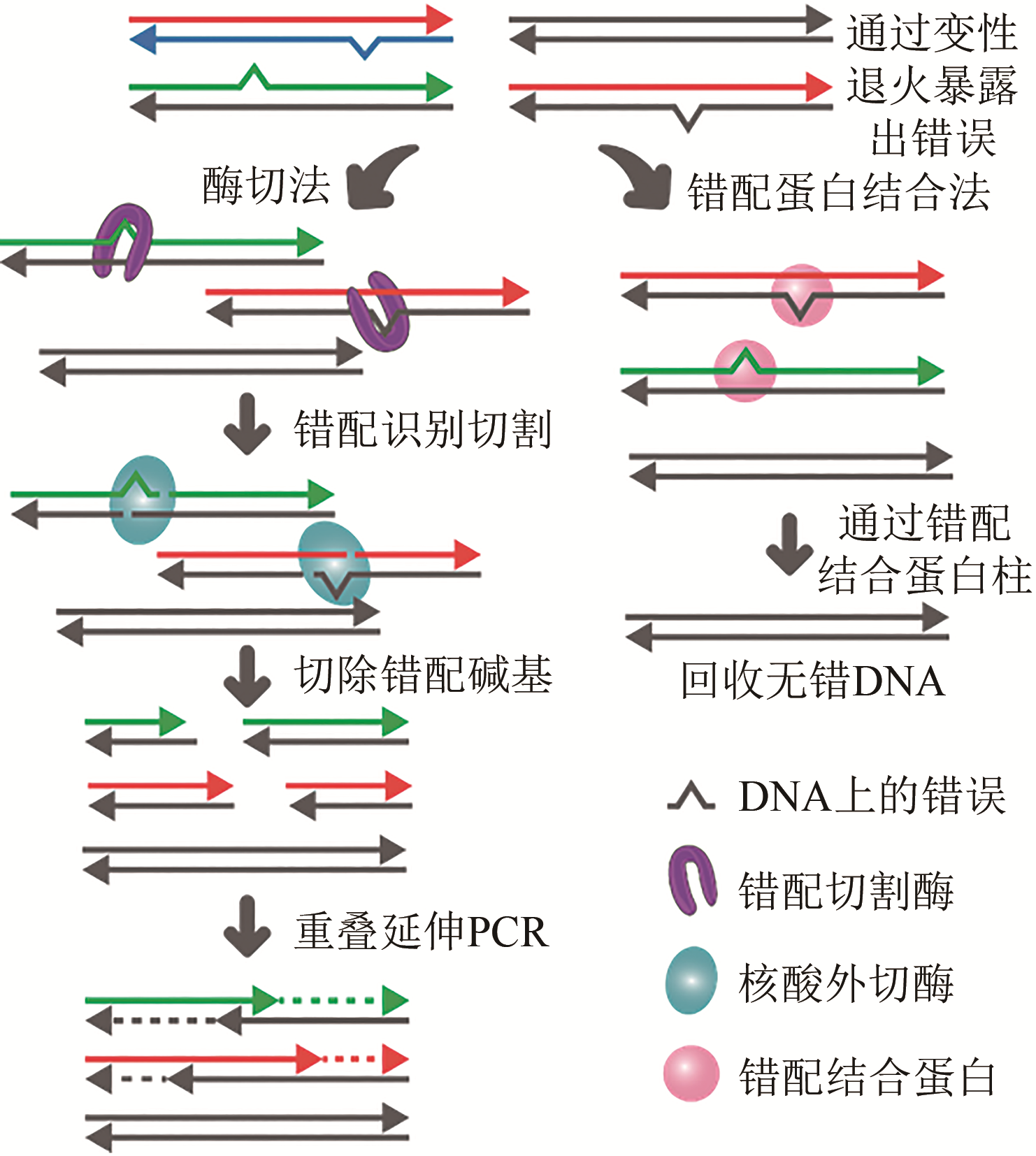
图4 基于错配切割和错配结合的纠错策略(Error rich DNA sequences are denatured and randomly re-hybridized to expose the errors introduced during oligo synthesis and subsequent assembly as bulging mismatches. The mismatches are then removed by error correction strategies using either mismatch-cleaving enzymes or mismatch-binding proteins. Fragments with fewer errors are recovered by overlap assembly extension PCR or filtering)
Fig. 4 Strategies for error-removal based on mismatch cleavage and mismatch binding
| 1 | GIBSON D G. Programming biological operating systems: genome design, assembly and activation[J]. Nature Methods, 2014, 11(5): 521-526. |
| 2 | CARROLL D. Genome engineering with targetable nucleases[J]. Annual Review Biochemistry, 2014, 83: 409-439. |
| 3 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 4 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age[J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 5 | ENDY D. Foundations for engineering biology[J]. Nature, 2005, 438(7067): 449-453. |
| 6 | MA S, TANG N, TIAN J. DNA synthesis, assembly and applications in synthetic biology[J]. Current Opinion in Chemical Biology, 2012, 16(3/4): 260-267. |
| 7 | CARUTHERS M H. The chemical synthesis of DNA/RNA: our gift to science[J]. Journal of Biological Chemistry, 2013, 288(2): 1420-1427. |
| 8 | PALLUK S, ARLOW D H, DE ROND T, et al. De novo DNA synthesis using polymerase-nucleotide conjugates[J]. Nature Biotechnology, 2018, 36(7): 645-650. |
| 9 | CZAR M J, ANDERSON J C, BADER J S, et al. Gene synthesis demystified[J]. Trends in Biotechnology, 2009, 27(2): 63-72. |
| 10 | MA S, SAAEM I, TIAN J. Error correction in gene synthesis technology[J]. Trends in Biotechnology, 2012, 30(3): 147-154. |
| 11 | BEAUCAGE S L, CARUTHERS M H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis[J]. Tetrahedron Letters, 1981, 22(20): 1859-1862. |
| 12 | TIAN J, MA K, SAAEM I. Advancing high-throughput gene synthesis technology[J]. Molecular Biosystems, 2009, 5(7): 714-722. |
| 13 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications[J]. Nature Methods, 2014, 11(5): 499-507. |
| 14 | LEPROUST E M, PECK B J, SPIRIN K, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process[J]. Nucleic Acids Research, 2010, 38(8): 2522-2540. |
| 15 | JENSEN Michael, ROBERTS Lester, JOHNSON Andrew, et al. Next generation 1536-well oligonucleotide synthesizer with on-the-fly dispense[J]. Journal of biotechnology, 2014, 171: 76-81. |
| 16 | FODOR S P, READ J L, PIRRUNG M C,et al. Light-directed, spatially addressable parallel chemical synthesis[J]. Science, 1991, 251(4995): 767-773. |
| 17 | BARONE A D, BEECHER J E, BURY P A, et al. Photolithographic synthesis of high-density oligonucleotide probe arrays[J]. Nucleosides Nucleic Acids, 2001, 20(4-7): 525-531. |
| 18 | SINGH-GASSON S, GREEN R D, YUE Y, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array[J]. Nature Biotechnology, 1999, 17(10): 974-978. |
| 19 | GAO X, LEPROUST E, ZHANG H, et al. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids[J]. Nucleic Acids Research, 2001, 29(22): 4744-4750. |
| 20 | AGBAVWE C, KIM C, HONG D, et al. Efficiency, error and yield in light-directed maskless synthesis of DNA microarrays[J]. Journal of Nanobiotechnology, 2011, 9: 57. |
| 21 | ROTH Kristian M, PEYVAN Kia, SCHWARZKOPF Kevin R, et al. Electrochemical detection of short DNA oligomer hybridization using the CombiMatrix ElectraSense microarray reader[J]. Electroanalysis, 2006, 18(19‐20): 1982-1988. |
| 22 | HUGHES T R, MAO M, JONES A R, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer[J]. Nature Biotechnology, 2001, 19(4): 342-347. |
| 23 | LIPSHUTZ R J, FODOR S P, GINGERAS T R, et al. High density synthetic oligonucleotide arrays[J]. Nature Genetics, 1999, 21(S1): 20-24. |
| 24 | MINHAZ UD-DEAN S M. A theoretical model for template-free synthesis of long DNA sequence[J]. Systems and Synthetic Biology, 2008, 2(3/4): 67-73. |
| 25 | MACKEY J K, GILHAM P T. New approach to the synthesis of polyribonucleotides of defined sequence[J]. Nature, 1971, 233(5321): 551-553. |
| 26 | GILLAM S, WATERMAN K, DOEL M, et al. Enzymatic synthesis of deoxyribo-oligonucleotides of defined sequence. Deoxyribo-oligonucleotide synthesis[J]. Nucleic Acids Research, 1974, 1(12): 1649-1664. |
| 27 | ENGLAND T E, UHLENBECK O C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase[J]. Biochemistry, 1978, 17(11): 2069-2076. |
| 28 | SCHMITZ Carole, REETZ Manfred T. Solid-phase enzymatic synthesis of oligonucleotides[J]. Organic Letters, 1999, 1(11): 1729-1731. |
| 29 | JENSEN M A, DAVIS R W. Template-independent enzymatic oligonucleotide synthesis (TiEOS): its history, prospects, and challenges[J]. Biochemistry, 2018, 57(12): 1821-1832. |
| 30 | BOLLUM F J. Thermal conversion of nonpriming deoxyribonucleic acid to primer [J]. The Journal of Biological Chemistry, 1959, 234(10): 2733-2734. |
| 31 | BOLLUM F J. Oligodeoxyribonucleotide-primed reactions catalyzed by calf thymus polymerase[J]. Journal of Biological Chemistry, 1962, 237: 1945-1949. |
| 32 | SCHOTT H, SCHRADE H. Single-step elongation of oligodeoxynucleotides using terminal deoxynucleotidyl transferase[J]. European Journal of Biochemistry, 1984, 143(3): 613-620. |
| 33 | TJONG V, YU H, HUCKNALL A, et al. Amplified on-chip fluorescence detection of DNA hybridization by surface-initiated enzymatic polymerization[J]. Analytical Chemistry, 2011, 83(13): 5153-5159. |
| 34 | MOTEA E A, BERDIS A J. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase[J]. Biochimica et Biophysica Acta(BBA) - Proteins and Proteomics, 2010, 1804(5): 1151-1166. |
| 35 | WU J, ZHANG S, MENG Q, et al. 3'-O-modified nucleotides as reversible terminators for pyrosequencing[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(42): 16462-16467. |
| 36 | KONG D S, CARR P A, CHEN L, et al. Parallel gene synthesis in a microfluidic device[J]. Nucleic Acids Research, 2007, 35(8): e61. |
| 37 | TIAN J, GONG H, SHENG N, et al. Accurate multiplex gene synthesis from programmable DNA microchips[J]. Nature, 2004, 432(7020): 1050-1054. |
| 38 | QUAN J, SAAEM I, TANG N, et al. Parallel on-chip gene synthesis and application to optimization of protein expression[J]. Nature Biotechnology, 2011, 29(5): 449-452. |
| 39 | ENGLER C, KANDZIA R, MARILLONNET S. A one pot, one step, precision cloning method with high throughput capability[J]. PLoS One, 2008, 3(11): e3647. |
| 40 | LI M Z, ELLEDGE S J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC[J]. Nature Methods, 2007, 4(3): 251-256. |
| 41 | ZHANG Y, WERLING U, EDELMANN W. SLiCE: a novel bacterial cell extract-based DNA cloning method[J]. Nucleic Acids Research, 2012, 40(8): e55. |
| 42 | Stefan de KOK, STANTON Leslie H, SLABY Todd, et al. Rapid and reliable DNA assembly via ligase cycling reaction[J]. ACS Synthetic Biology, 2014, 3(2): 97-106. |
| 43 | QUAN J, TIAN J. Circular polymerase extension cloning of complex gene libraries and pathways[J]. PLoS One, 2009, 4(7): e6441. |
| 44 | GIBSON D G, YOUNG L, CHUANG R Y, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nature Methods, 2009, 6(5): 343-345. |
| 45 | JUHAS M, AJIOKA J W. High molecular weight DNA assembly in vivo for synthetic biology applications[J]. Critical Reviews in Biotechnology, 2017, 37(3): 277-286. |
| 46 | SHIZUYA H, BIRREN B, KIM U J, et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector[J]. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(18): 8794-8797. |
| 47 | KANEKO S, TSUGE K, TAKEUCHI T, et al. Conversion of sub-megasized DNA to desired structures using a novel Bacillus subtilis genome vector[J]. Nucleic Acids Research, 2003, 31(18): e112. |
| 48 | BURKE D T, CARLE G F, OLSON M V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors[J]. Science, 1987, 236(4803): 806-812. |
| 49 | OGAWA T, IWATA T, KANEKO S, et al. An inducible recA expression Bacillus subtilis genome vector for stable manipulation of large DNA fragments[J]. Biotechnology & Applied Microbiology Genomics, 2015, 16(1): 209. |
| 50 | GIBSON D G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides[J]. Nucleic Acids Research, 2009, 37(20): 6984-6990. |
| 51 | LIN Q, JIA B, MITCHELL L A, et al. RADOM, an efficient in vivo method for assembling designed DNA fragments up to 10 kb long in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2015, 4(3): 213-220. |
| 52 | JAKOCIUNAS T, RAJKUMAR A S, ZHANG J, et al. CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2015, 4(11): 1226-1234. |
| 53 | GIBSON D G, BENDERS G A, AXELROD K C, et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(51): 20404-20409. |
| 54 | SHAO Y, LU N, WU Z, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 55 | LUBOCK N B, ZHANG D, SIDORE A M, et al. A systematic comparison of error correction enzymes by next-generation sequencing[J]. Nucleic Acids Research, 2017, 45(15): 9206-9217. |
| 56 | SINHA N D, JUNG K E. Analysis and purification of synthetic nucleic acids using HPLC[J]. Current Protocols in Nucleic Acid Chemistry, 2015. DOI: 10.1002/0471142700.nc1005s61 . |
| 57 | ELLINGTON A, POLLARD J D, Jr. Introduction to the synthesis and purification of oligonucleotides[J]. Current Protocols in Nucleic Acid Chemistry, 2000. DOI: 10.1002/0471142700. nca03cs00 . |
| 58 | BOROVKOV A Y, LOSKUTOV A V, ROBIDA M D, et al. High-quality gene assembly directly from unpurified mixtures of microarray-synthesized oligonucleotides[J]. Nucleic Acids Research, 2010, 38(19): e180. |
| 59 | MATZAS M, STAHLER P F, KEFER N, et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing[J]. Nature Biotechnology, 2010, 28(12): 1291-1294. |
| 60 | SANCAR A, LINDSEY-BOLTZ L A, UNSAL-KACMAZ K, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints[J]. Annual Review of Biochemistry, 2004, 73: 39-85. |
| 61 | JIRICNY J. The multifaceted mismatch-repair system[J]. Nature Reviews Molecular Cell Biology, 2006, 7(5): 335-346. |
| 62 | LEE J B, CHO W K, PARK J, et al. Single-molecule views of MutS on mismatched DNA[J]. DNA Repair (Amst), 2014, 20: 82-93. |
| 63 | KUNKEL T A, ERIE D A. DNA mismatch repair[J]. Annual Review of Biochemistry, 2005, 74: 681-710. |
| 64 | CARR P A, PARK J S, LEE Y J, et al. Protein-mediated error correction for de novo DNA synthesis[J]. Nucleic Acids Research, 2004, 32(20): e162. |
| 65 | WAN W, LI L, XU Q, et al. Error removal in microchip-synthesized DNA using immobilized MutS[J]. Nucleic Acids Research, 2014, 42(12): e102. |
| 66 | ZHANG J, WANG Y, CHAI B, et al. Efficient and low-cost error removal in DNA synthesis by a high-durability MutS[J]. ACS Synthetic Biology, 2020, 9(4): 940-952. |
| 67 | BINKOWSKI B F, RICHMOND K E, KAYSEN J, et al. Correcting errors in synthetic DNA through consensus shuffling[J]. Nucleic Acids Research, 2005, 33(6): e55. |
| 68 | TILL B J, BURTNER C, COMAI L, et al. Mismatch cleavage by single-strand specific nucleases[J]. Nucleic Acids Research, 2004, 32(8): 2632-2641. |
| 69 | FUHRMANN M. Removal of mismatched bases from synthetic genes by enzymatic mismatch cleavage[J]. Nucleic Acids Research, 2005, 33(6): e58. |
| 70 | DESAI N A, SHANKAR V. Single-strand-specific nucleases[J]. FEMS Microbiology Reviews, 2003, 26(5): 457-491. |
| 71 | SEQUEIRA A F, GUERREIRO C I, VINCENTELLI R, et al. T7 Endonuclease I mediates error correction in artificial gene synthesis[J]. Molecular Biotechnology, 2016, 58(8-9): 573-584. |
| 72 | BANG D, CHURCH G M. Gene synthesis by circular assembly amplification[J]. Nature Methods, 2008, 5(1): 37-39. |
| 73 | BABON J J, MCKENZIE M, COTTON R G. Mutation detection using fluorescent enzyme mismatch cleavage with T4 endonuclease Ⅶ[J]. Electrophoresis, 1999, 20(6): 1162-1170. |
| 74 | YEUNG A T, HATTANGADI D, BLAKESLEY L, et al. Enzymatic mutation detection technologies[J]. Biotechniques, 2005, 38(5): 749-758. |
| 75 | OLEYKOWSKI C A, BRONSON MULLINS C R, GODWIN A K, et al. Mutation detection using a novel plant endonuclease[J]. Nucleic Acids Research, 1998, 26(20): 4597-4602. |
| 76 | YANG B, WEN X, KODALI N S, et al. Purification, cloning, and characterization of the CEL I nuclease[J]. Biochemistry, 2000, 39(13): 3533-3541. |
| 77 | SAAEM I, MA S, QUAN J, et al. Error correction of microchip synthesized genes using Surveyor nuclease[J]. Nucleic Acids Research, 2012, 40(3): e23. |
| 78 | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synⅡ, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355(6329) : eaaf4791. |
| 79 | WU Y, LI B Z, ZHAO M,et al. Bug mapping and fitness testing of chemically synthesized chromosome Ⅹ[J]. Science, 2017, 355(6329) : eaaf4706. |
| 80 | XIE Z X, LI B Z, MITCHELL L A, et al. ‘Perfect’ designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329) : eaaf4704. |
| 81 | ZHANG W, ZHAO G, LUO Z, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355(6329) : eaaf3981. |
| 82 | RICHARDSON S M, MITCHELL L A, STRACQUADANIO G., et al. Design of a synthetic yeast genome[J]. Science, 2017, 355(6329): 1040-1044. |
| 83 | SEEMAN Nadrian C, SLEIMAN Hanadi F. DNA nanotechnology[J]. Nature Reviews Materials, 2017, 3(1) :17068. |
| 84 | GOLDMAN N, BERTONE P, CHEN S, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA[J]. Nature, 2013, 494(7435): 77-80. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 张宣梁, 李青婷, 王飞. DNA存储系统中的数据写入[J]. 合成生物学, 2024, 5(5): 1125-1141. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
