合成生物学 ›› 2021, Vol. 2 ›› Issue (3): 309-322.DOI: 10.12211/2096-8280.2021-001
DNA信息存储:生命系统与信息系统的桥梁
韩明哲1,2, 陈为刚1,3, 宋理富1,2, 李炳志1,2, 元英进1,2
- 1.天津大学,合成生物学前沿科学中心,系统生物工程教育部重点实验室,天津 300072
2.天津大学化工学院,天津 300072
3.天津大学微电子学院,天津 300072
-
收稿日期:2021-01-04修回日期:2021-03-02出版日期:2021-06-30发布日期:2021-07-13 -
通讯作者:元英进 -
作者简介:韩明哲 (1996—),男,博士研究生。研究方向为合成生物学及DNA信息存储。E-mail:mickeyhan@tju.edu.cn元英进 (1963—),男,教授,博士生导师。研究方向为合成生物学及人工基因组化学合成。E-mail:yjyuan@tju.edu.cn
DNA information storage: bridging biological and digital world
HAN Mingzhe1,2, CHEN Weigang1,3, SONG Lifu1,2, LI Bingzhi1,2, YUAN Yingjin1,2
- 1.Frontier Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education),Tianjin University,Tianjin 300072,China
2.School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
3.School of Microelectronics,Tianjin University,Tianjin 300072,China
-
Received:2021-01-04Revised:2021-03-02Online:2021-06-30Published:2021-07-13 -
Contact:YUAN Yingjin
摘要:
DNA信息存储通过编解码、合成、编辑和测序等过程,实现数字信息写入、存储与读出。其在密度、寿命、能耗和抗电磁干扰等方面较磁、光、电等常规的信息存储介质有较大优势。随着全球数据总量的快速增长,DNA信息存储的优势特性和发展潜力受到了研究者的广泛关注。本文阐述了DNA信息存储的基本原理和技术流程,分析了DNA信息存储与生命系统和信息系统的关联,并依据读写技术特点归纳近年来涌现的“DNA硬盘”“DNA光盘”“DNA磁带”等几种主要模式、发展现状及技术路线。在此基础上,探讨DNA信息存储商业化、大规模应用面临的主要挑战,讨论更低成本的数据写入和更快速的数据读出,并指出可行的发展路线。最后,展望了DNA作为新型存储介质在现代存储系统中的发展演化趋势。
中图分类号:
引用本文
韩明哲, 陈为刚, 宋理富, 李炳志, 元英进. DNA信息存储:生命系统与信息系统的桥梁[J]. 合成生物学, 2021, 2(3): 309-322.
HAN Mingzhe, CHEN Weigang, SONG Lifu, LI Bingzhi, YUAN Yingjin. DNA information storage: bridging biological and digital world[J]. Synthetic Biology Journal, 2021, 2(3): 309-322.
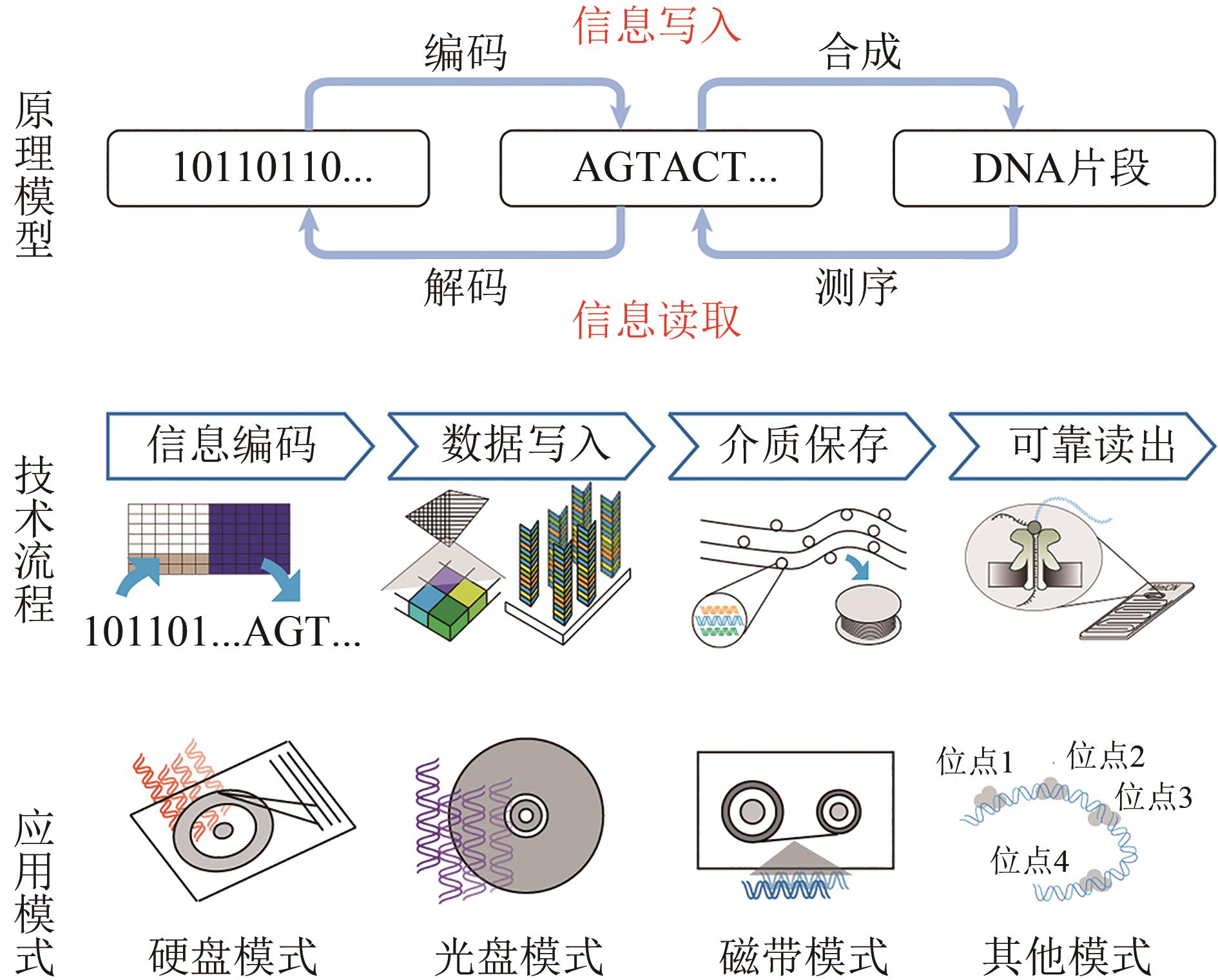
图1 DNA存储的原理模型、技术流程和应用模式(The basic principle is the conversion of digital information between binary code stream, quaternary base sequence and actual DNA fragment. The technical work flow includes Information encoding, data writing, media storage and reliable reading. Storage modes include "DNA hard drive", "DNA CD", "DNA tape" and others)
Fig. 1 The basic principle, technical work flow and storage modes of DNA information storage
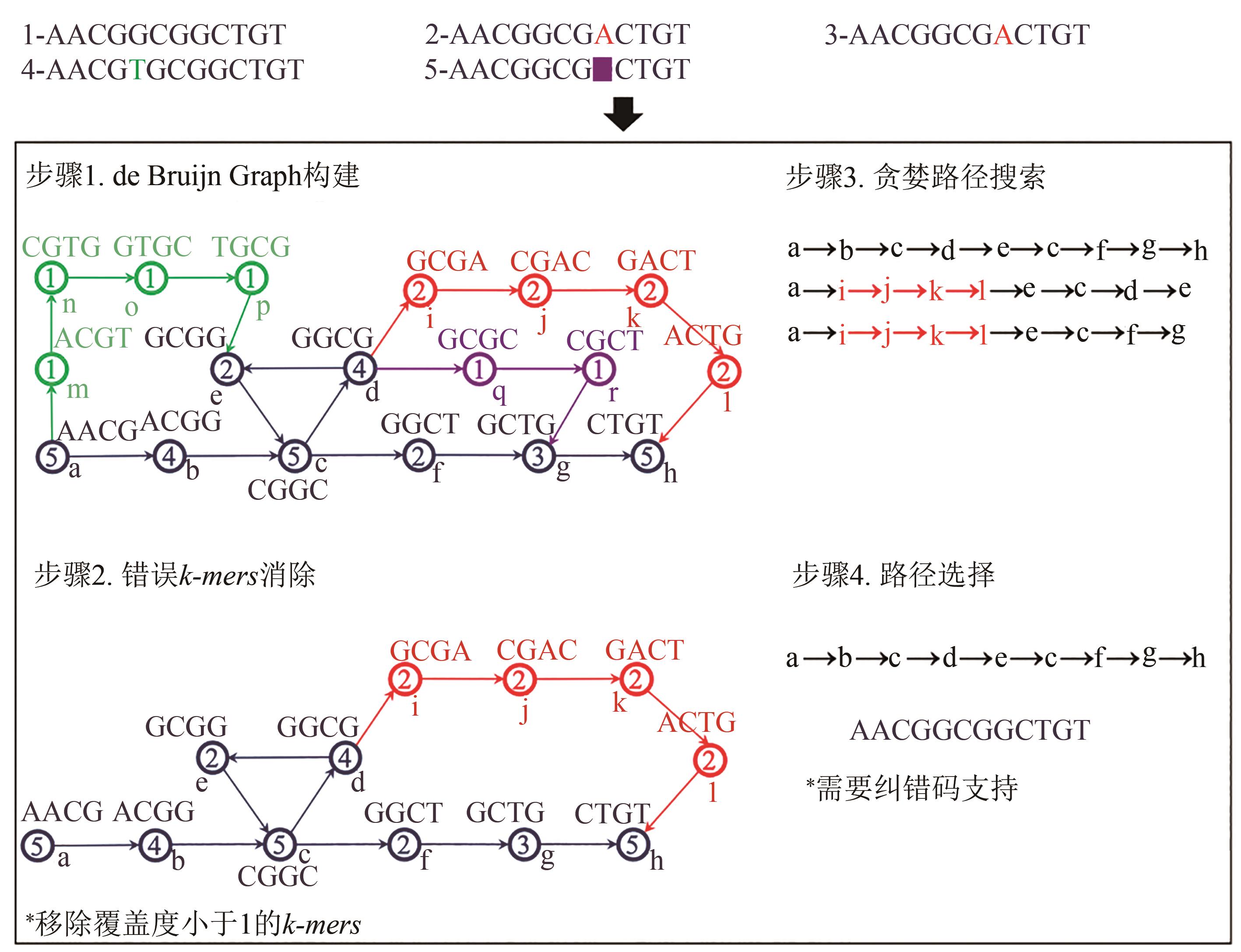
图2 基于de Bruijn图论的DNA序列重建算法[34](There are four major steps in this algorithm: Step 1—Construct the de Bruijn graph using the sequencing results; Step 2—Eliminate the noisy k-mers by removing the low coverage k-mers; Step 3—Greedy path search with the simplified de Bruijn graph to reveal all possible paths associated with specific indexes; Step 4—Select the correct paths, i.e. the correct strands, based on the embedded EC codes)
Fig. 2 Algorithm of de Bruijn graph-based reconstruction of DNA strands[34]
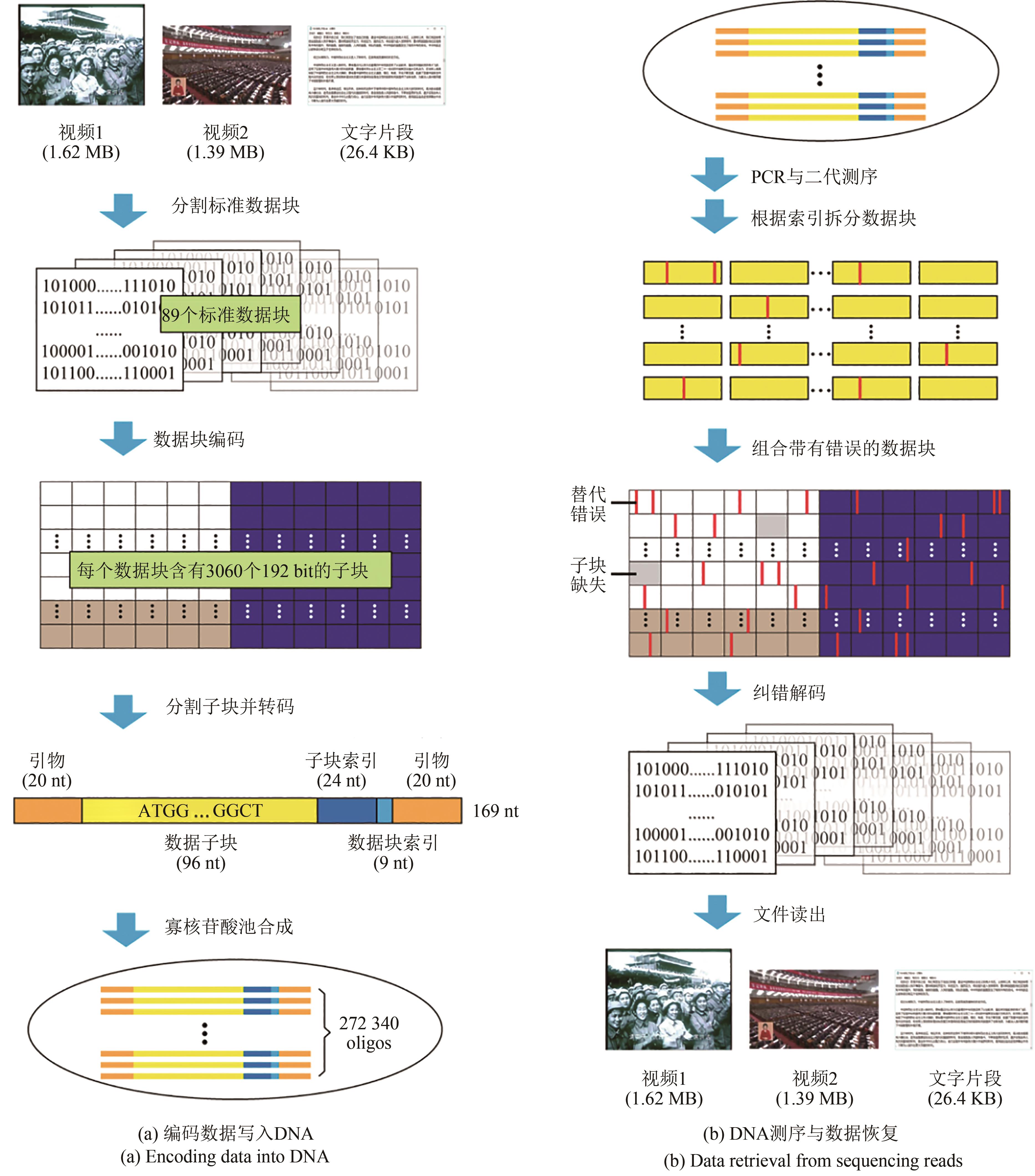
图3 “DNA硬盘”模式示意图[44][(a) Encoding data into DNA. The original files were converted into 89 standard data blocks and then divided into sub-blocks after error correction coding. Each 192-nt sub-block was transcoded into a 96-nt DNA sequence, and then the sub-block index, block index, and primers are added to form an oligonucleotide structure. (b) Data retrieval from sequencing reads. After PCR and 2nd-generation sequencing of synthesized oligo pool, data blocks with substitutions and erasures were obtained from sequencing reads according to indexes. With the help of error correction coding, original files were finally recovered]
Fig. 3 Schematic diagram of "DNA hard drive"[44]

图4 “DNA光盘”模式示意图[48][(a) Construction of the data-carrying chromosome. The DNA sequences encoded from original files were synthesized and in vivo assembled into a 254 886 bp chromosome. (b) Encoding scheme. The encoding process included LDPC coding, random interleaver, sparsification, superposition with watermark sequences, and transcoding into DNA sequences. (c) Data retrieval from nanopore readout. The data retrieval process included de novo assembly, polishing, locating, indel correction with watermark sequences, and substitution error correction with the LDPC code]
Fig. 4 Schematic diagram of "DNA CD"[48]

图5 DNA信息存储成本比较与预测(Blue spots, HDD; green spots, floppy disk; purple spots, RAM; orange spots, DNA information storage)
Fig. 5 Comparison and forecast of cost by DNA information storage
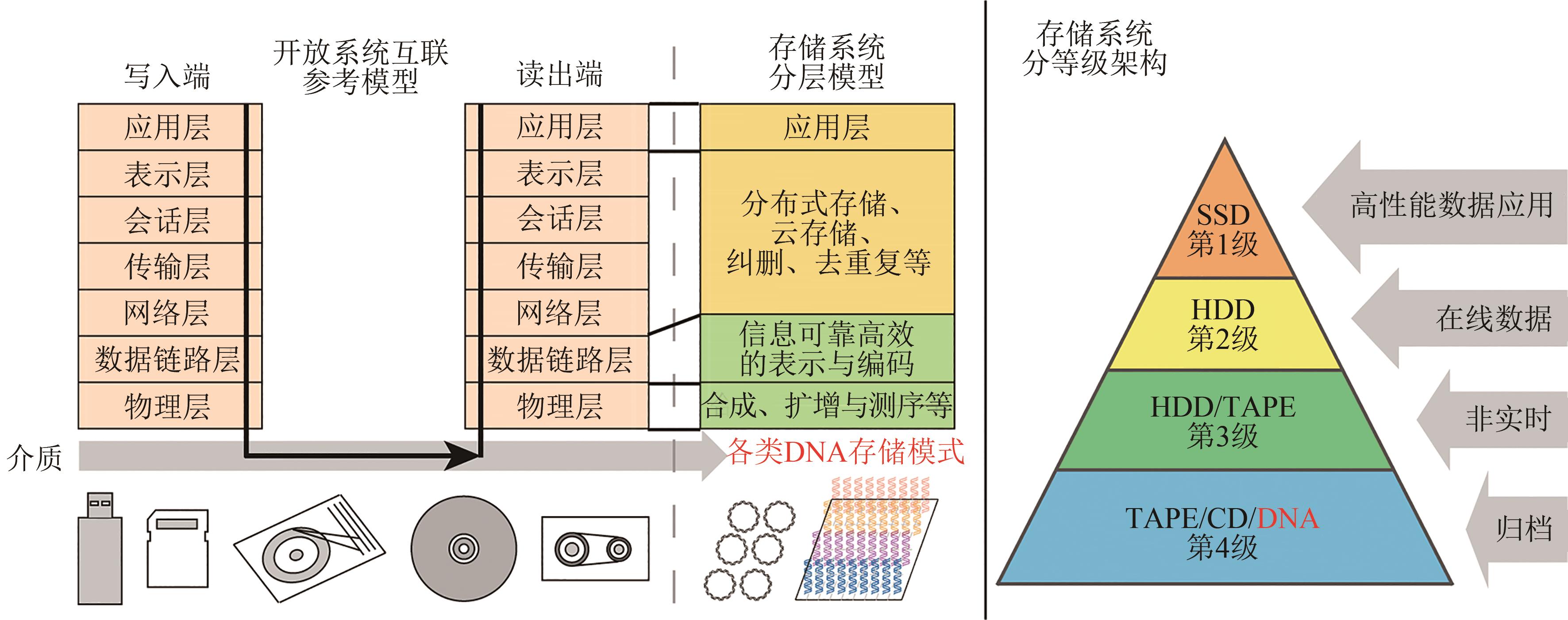
图7 DNA信息存储与现代存储系统的融合(Synthetic DNA enriches the modern data storage media including SSD, HDD, CD, tape. The introduced new property by DNA requires not only suited low-level techniques including synthesis, amplification and sequencing, but also reliability guarantee schemes such as error correction codes, erasure correction, redundancy eliminating schemes, etc. New application paradigms also should be explored to dig out this new medium. The new medium changes all the elements in the 7 levels of OSI reference model and reforms the classic storage hierarchical architecture)
Fig. 7 Fusion of DNA information storage and information storage system
| 1 | Semiconductor Industry Association. Decadal plan for semiconductors [EB/OL]. [2020-12-02]. . |
| 2 | FONTANA R E, DECAD G M, HETZLER S R. Volumetric density trends (TB/in.3) for storage components: TAPE, hard disk drives, NAND, and Blu-ray[J]. Journal of Applied Physics, 2015, 117(17): 17E301. |
| 3 | ZHIRNOV V, ZADEGAN R M, SANDHU G S, et al. Nucleic acid memory[J]. Nature Materials, 2016, 15(4): 366-370. |
| 4 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications[J]. Nature Methods, 2014, 11(5): 499-507. |
| 5 | WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): eaaf4706. |
| 6 | XIE Z X, LI B Z, MITCHELL L A, et al. "Perfect" designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 7 | 陈欣懋, 欧阳颀. 生物逆向工程设计在合成生物学中的应用[J]. 合成生物学, 2020, 1(1): 29-43. |
| CHEN X M, OUYANG Q. The application of biological reverse engineering in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 29-43. | |
| 8 | 彭凯, 逯晓云, 程健, 等. DNA合成、组装与纠错技术研究进展[J]. 合成生物学, 2020, 1(6): 697-708. |
| PENG K, LU X Y, CHENG J, et al. Advances in technologies for de novo DNA synthesis, assembly and error correction[J]. Synthetic Biology Journal, 2020, 1(6): 697-708. | |
| 9 | 曹中正, 张心怡, 徐艺源, 等. 基因组编辑技术及其在合成生物学中的应用[J]. 合成生物学, 2020, 1(4): 413-426. |
| CAO Z Z, ZHANG X Y, XU Y Y, et al. Genome editing technology and its applications in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(4): 413-426. | |
| 10 | 王会, 戴俊彪, 罗周卿. 基因组的"读-改-写"技术[J]. 合成生物学, 2020, 1(5): 503-515. |
| WANG H, DAI J B, LUO Z Q. Reading, editing, and writing techniques for genome research[J]. Synthetic Biology Journal, 2020, 1(5): 503-515. | |
| 11 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING M Z, LI B Z, WANG Y, et al. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 12 | Wire Business. Twist Bioscience, Illumina and Western Digital form alliance with Microsoft to advance data storage in DNA [EB/OL]. [2020-11-12]. . |
| 13 | CEZE L, NIVALA J, STRAUSS K. Molecular digital data storage using DNA[J]. Nature Reviews Genetics, 2019, 20(8): 456-66. |
| 14 | MEISER L C, ANTKOWIAK P L, KOCH J, et al. Reading and writing digital data in DNA[J]. Nature Protocols, 2020, 15(1): 86-101. |
| 15 | DONG Y, SUN F, PING Z, et al. DNA storage: research landscape and future prospects[J]. National Science Review, 2020, 7(6): 1092-1107. |
| 16 | PING Z, MA D, HUANG X, et al. Carbon-based archiving: current progress and future prospects of DNA-based data storage[J]. GigaScience, 2019, 8(6): giz075. |
| 17 | LIM C K, NIRANTAR S, YEW W S, et al. Novel modalities in DNA data storage[J]. Trends in Biotechnology, 2021. DOI: https://doi.org/10.1016/j.tibtech.2020.12.008 . |
| 18 | 周廷尧, 罗源, 蒋兴宇. DNA数据存储:保存策略与数据加密[J]. 合成生物学, 2021, 2(3): 371-383. |
| ZHOU T Y, LUO Y, JIANG X Y. DNA data storage: preservation approach and data encryption[J]. Synthetic Biology Journal, 2021, 2(3): 371-383. | |
| 19 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA[J]. Science, 2012, 337(6102): 1628. |
| 20 | KEIGER D. DNA hard drive [EB/OL]. [2012-11-30]. . |
| 21 | 毕昆, 顾万君, 陆祖宏. DNA 存储中的编码技术[J]. 生物信息学, 2020, 18(2): 76-85. |
| BI K, GU W J, LU Z H. Coding algorithms in DNA storage[J]. Chinese Journal of Bioinformatics, 2020, 18(2): 76-85. | |
| 22 | Bioscience Twist. Product sheet of Twist oligo pools [EB/OL]. [2019-08-29]. |
| 23 | GenScript. Precise synthetic oligo pools [EB/OL]. [2021-02-01]. . |
| 24 | LIU L, LI Y, LI S, et al. Comparison of next-generation sequencing systems [J]. Journal of Biomedicine and Biotechnology, 2012, 2012: 251364. |
| 25 | GOLDMAN N, BERTONE P, CHEN S Y, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA [J]. Nature, 2013, 494(7435): 77-80. |
| 26 | GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes[J]. Angewandte Chemie International Edition, 2015, 54(8): 2552-2555. |
| 27 | ERLICH Y, ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture[J]. Science, 2017, 355(6328): 950-953. |
| 28 | ANAVY L, VAKNIN I, ATAR O, et al. Data storage in DNA with fewer synthesis cycles using composite DNA letters[J]. Nature Biotechnology, 2019, 37(10): 1229-1236. |
| 29 | CHOI Y, RYU T, LEE A C, et al. High information capacity DNA-based data storage with augmented encoding characters using degenerate bases[J]. Scientific Reports, 2019, 9(1): 6582. |
| 30 | HOSHIKA S, LEAL N A, KIM M J, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks[J]. Science, 2019, 363(6429): 884-887. |
| 31 | HECKEL R, SHOMORONY I, RAMCHANDRAN K, et al. Fundamental limits of DNA storage systems[C]//2017 IEEE International Symposium on Information Theory (ISIT). Aachen, Germany: IEEE, 2017: 3130-3134. |
| 32 | PRESS W H, HAWKINS J A, JONES S K, et al. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(31): 18489-18496. |
| 33 | SABARY O, YUCOVICH A, SHAPIRA G, et al. Reconstruction Algorithms for DNA-Storage Systems[EB/OL]. [2021-05-25]. . |
| 34 | SONG L, GENG F, GONG Z, et al. Super-robust data storage in DNA by de Bruijn graph-based decoding[EB/OL]. [2021-05-25]. . |
| 35 | LEE H H, KALHOR R, GOELA N, et al. Terminator-free template-independent enzymatic DNA synthesis for digital information storage[J]. Nature Communications, 2019, 10(1): 2383. |
| 36 | LEE H, WIEGAND D J, GRISWOLD K, et al. Photon-directed multiplexed enzymatic DNA synthesis for molecular digital data storage[J]. Nature Communications, 2020, 11(1): 5246. |
| 37 | LIN K N, VOLKEL K, TUCK J M, et al. Dynamic and scalable DNA-based information storage[J]. Nature Communications, 2020, 11(1), 2981. |
| 38 | CHOI Y, BAE H J, LEE A C, et al. DNA micro‐disks for the management of DNA‐based data storage with Index and Write‐Once-Read‐Many (WORM) memory features[J]. Advanced Materials, 2020, 32(37): 2001249. |
| 39 | GAO Y, CHEN X, QIAO H, et al. Low-bias manipulation of DNA oligo pool for robust data storage[J]. ACS Synthetic Biology, 2020, 9(12): 3344-3352. |
| 40 | ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36(7): 242-248. |
| 41 | TAKAHASHI C N, NGUYEN B H, STRAUSS K, et al. Demonstration of end-to-end automation of DNA data storage[J]. Scientific Reports, 2019, 9(1): 4998. |
| 42 | NEWMAN S, STEPHENSON A P, WILLSEY M, et al. High density DNA data storage library via dehydration with digital microfluidic retrieval[J]. Nature Communications, 2019, 10(1): 1706. |
| 43 | SHANKLAND S. Startup packs all 16GB of Wikipedia onto DNA strands to demonstrate new storage tech [EB/OL]. [2019-07-02]. . |
| 44 | 陈为刚, 黄刚, 李炳志, 等. 音视频文件的DNA信息存储[J]. 中国科学:生命科学, 2019, 50(1): 81-85. |
| CHEN W G, HUANG G, LI B Z, et al. DNA information storage for audio and video files[J]. SCIENTIA SINICA Vitae, 2019, 50(1): 81-85. | |
| 45 | PING Z, CHEN S, ZHOU G, et al. Towards practical and robust DNA-based data archiving by codec system named 'Yin-Yang'[EB/OL]. [2021-05-25]. . |
| 46 | BORNHOLT J, LOPEZ R, CARMEAN D M, et al. Toward a DNA-based archival storage system[J]. IEEE Micro, 2017, 37(3): 98-104. |
| 47 | YAZDI S M H T, YUAN Y, MA J, et al. A rewritable, random-access DNA-based storage system[J]. Scientific Reports, 2015, 5(1): 14138. |
| 48 | CHEN W G, HAN M Z, ZHOU J T, et al. An artificial chromosome for data storage[J]. National Science Review, 2021, 8(5): nwab028. |
| 49 | ZHU Y O, SIEGAl M L, HALL D W, et al. Precise estimates of mutation rate and spectrum in yeast[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(22): E2310-E2318. |
| 50 | LEE H, POPODI E, TANG H, et al. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(41): E2774-E2783. |
| 51 | SONG L F, ZENG A P. Orthogonal information encoding in living cells with high error- tolerance, safety, and fidelity [J]. ACS Synthetic Biology, 2018, 7(3): 866-874. |
| 52 | DEAMER D, AKESON M, BRANTON D. Three decades of nanopore sequencing[J]. Nature Biotechnology, 2016, 34(5): 518-524. |
| 53 | DAVIS J. Microvenus[J]. Art Journal, 1996, 55(1): 70-74. |
| 54 | BANCROFT C, BOWLER T, BLOOM B, et al. Long-term storage of information in DNA[J]. Science, 2001, 293(5536): 1763-1765. |
| 55 | WONG P C, WONG K K, FOOTE H. Organic data memory using the DNA approach[J]. Communications of the ACM, 2003, 46(1): 95-98. |
| 56 | GUSTAFSSON C. For anyone who ever said there's no such thing as a poetic gene[J]. Nature, 2009, 458(7239): 703. |
| 57 | YACHIE N, SEKIYAMA K, SUGAHARA J, et al. Alignment-based approach for durable data storage into living organisms[J]. Biotechnology Progress, 2007, 23(2): 501-505. |
| 58 | AILENBERG M, ROTSTEIN O D. An improved Huffman coding method for archiving text, images, and music characters in DNA[J]. Biotechniques, 2009, 47(3): 747-751. |
| 59 | NGUYEN H H, PARK J, PARK S J, et al. Long-term stability and integrity of plasmid-based DNA data storage[J]. Polymers, 2018, 10(1): 28. |
| 60 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 61 | SHIPMAN S L, NIVALA J, MACKLIS J D, et al. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria[J]. Nature, 2017, 547(7663): 345-349. |
| 62 | HAO M, QIAO H, GAO Y, et al. A mixed culture of bacterial cells enables an economic DNA storage on a large scale[J]. Communications Biology, 2020, 3(1): 416. |
| 63 | AUSLÄNDER S, FUSSENEGGER M. Dynamic genome engineering in living cells[J]. Science, 2014, 346(6211): 813-814. |
| 64 | FARZADFARD F, LU T K. Emerging applications for DNA writers and molecular recorders[J]. Science, 2018, 361(6405): 870-875. |
| 65 | FARZADFARD F, LU T K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations[J]. Science, 2014, 346(6211): . |
| 66 | SHETH R U, YIM S S, WU F L, et al. Multiplex recording of cellular events over time on CRISPR biological tape[J]. Science, 2017, 358(6369): 1457-1461. |
| 67 | TANG W, LIU D R. Rewritable multi-event analog recording in bacterial and mammalian cells[J]. Science, 2018, 360(6385): eaap8992. |
| 68 | PERLI S D, CUI C H, LU T K. Continuous genetic recording with self-targeting CRISPR-Cas in human cells[J]. Science, 2016, 353(6304): aag0511. |
| 69 | FARZADFARD F, GHARAEI N, HIGASHIKUNI Y, et al. Single-nucleotide-resolution computing and memory in living cells[J]. Molecular Cell, 2019, 75(4): 769-780. |
| 70 | YIM S S, MCBEE R M, SONG A M, et al. Robust direct digital-to-biological data storage in living cells[J]. Nature Chemical Biology, 2021, 17(3):246-253. |
| 71 | TABATABAEI S K, WANG B, ATHREYA N B M, et al. DNA punch cards for storing data on native DNA sequences via enzymatic nicking[J]. Nature communications, 2020, 11(1): 1742. |
| 72 | CHEN K, KONG J, ZHU J, et al. Digital data storage using DNA nanostructures and solid-state nanopores[J]. Nano Letters, 2018, 19(2): 1210-1215. |
| 73 | CHEN K, ZHU J, BOSKOVIC F, et al. Nanopore-based DNA hard drives for rewritable and secure data storage[J]. Nano Letters, 2020, 20(5): 3754-3760. |
| 74 | LOPEZ R, CHEN Y J, ANG S D, et al. DNA assembly for nanopore data storage readout[J]. Nature Communications, 2019, 10(1): 2933. |
| 75 | ZHANG Y, WANG F, CHAO J, et al. DNA origami cryptography for secure communication[J]. Nature Communications, 2019, 10(1): 5469. |
| 76 | Semiconductor Research Corporation. 2018 semiconductor synthetic biology roadmap [R]. Durham, NC, USA: SRC, 2018. |
| 77 | MCCALLUM J C. Disk drive prices 1955+ [EB/OL]. [2021-05-25]. . |
| 78 | IARPA. IARPA BAA on molecular information storage [EB/OL]. [2021-05-25]. . |
| 79 | MARKOWITZ D. SRC/IARPA Workshop on DNA-based massive information storage [EB/OL]. [2021-05-25]. . |
| 80 | ANTKOWIAK P L, LIETARD J, DARESTANI M Z, et al. Low cost DNA data storage using photolithographic synthesis and advanced information reconstruction and error correction[J]. Nature Communications, 2020, 11(1): 5345. |
| 81 | Illumina. Illumina sequencing platforms [EB/OL]. [2021-05-25]. |
| 82 | Illumina. Run time estimates for each sequencing step on the Illumina sequencing platforms. [EB/OL]. [2021-05-25]. . |
| 83 | KONO N, ARAKAWA K. Nanopore sequencing: review of potential applications in functional genomics[J]. Development, Growth & Differentiation, 2019, 61(5): 316-326. |
| 84 | YUAN Z, LIU Y, DAI M, et al. Controlling DNA translocation through solid-state nanopores[J]. Nanoscale Research Letters, 2020, 15: 80. |
| 85 | STANLEY P M, STRITTMATTER L M, VICKERS A M, et al. Decoding DNA data storage for investment[J]. Biotechnology Advances, 2020, 45: 107639. |
| 86 | HECKEL R, MIKUTIS G, GRASS R N. A characterization of the DNA data storage channel[J]. Scientific Reports, 2019, 9(1): 9663. |
| 87 | MAO W, DIGGAVI S N, KANNAN S. Models and information-theoretic bounds for nanopore sequencing[J]. IEEE Transactions on Information Theory, 2018, 64(4): 3216-3236. |
| 88 | MERCIER H, BHARGAVA V K, TAROKH V. A survey of error-correcting codes for channels with symbol synchronization errors[J]. IEEE Communications Surveys & Tutorials, 2010, 12(1): 87-96. |
| 89 | 平质, 张颢龄, 陈世宏, 等. Chamaeleo: DNA存储碱基编解码算法的可拓展集成与系统评估平台[J]. 合成生物学, 2021, 2(3): 412-427. |
| PING Z, ZHANG H L, CHEN S H, et al. Chamaeleo: an integrated evaluation platform for DNA storage[J]. Synthetic Biology Journal, 2021, 2(3): 412-427. | |
| 90 | Semiconductor Research Corporation. Decadal plan for semiconductors full report [R]. Durham: SRC, 2021. |
| 91 | PUZIS R, FARBIASH D, BRODT O, et al. Increased cyber-biosecurity for DNA synthesis [J]. Nature Biotechnology, 2020, 38(12): 1379-1381. |
| 92 | NEY P, KOSCHER K, ORGANICK L, et al. Computer security, privacy, and DNA sequencing: compromising computers with synthesized DNA, privacy leaks, and more[C]//26th USENIX Security Symposium (USENIX Security 17). Vancouver, BC, Canada: USENIX Association, 2017: 765-779. |
| 93 | KOCH J, GANTENBEIN S, MASANIA K, et al. A DNA-of-things storage architecture to create materials with embedded memory [J]. Nature Biotechnology, 2020, 38(1): 39-43. |
| 94 | Gartner. Gartner unveils top predictions for IT organizations and users in 2021 and beyond [EB/OL]. Gartner [2020-10-21]. . |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 张宣梁, 李青婷, 王飞. DNA存储系统中的数据写入[J]. 合成生物学, 2024, 5(5): 1125-1141. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||