Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (1): 1-15.DOI: 10.12211/2096-8280.2023-038
• Invited Review • Previous Articles Next Articles
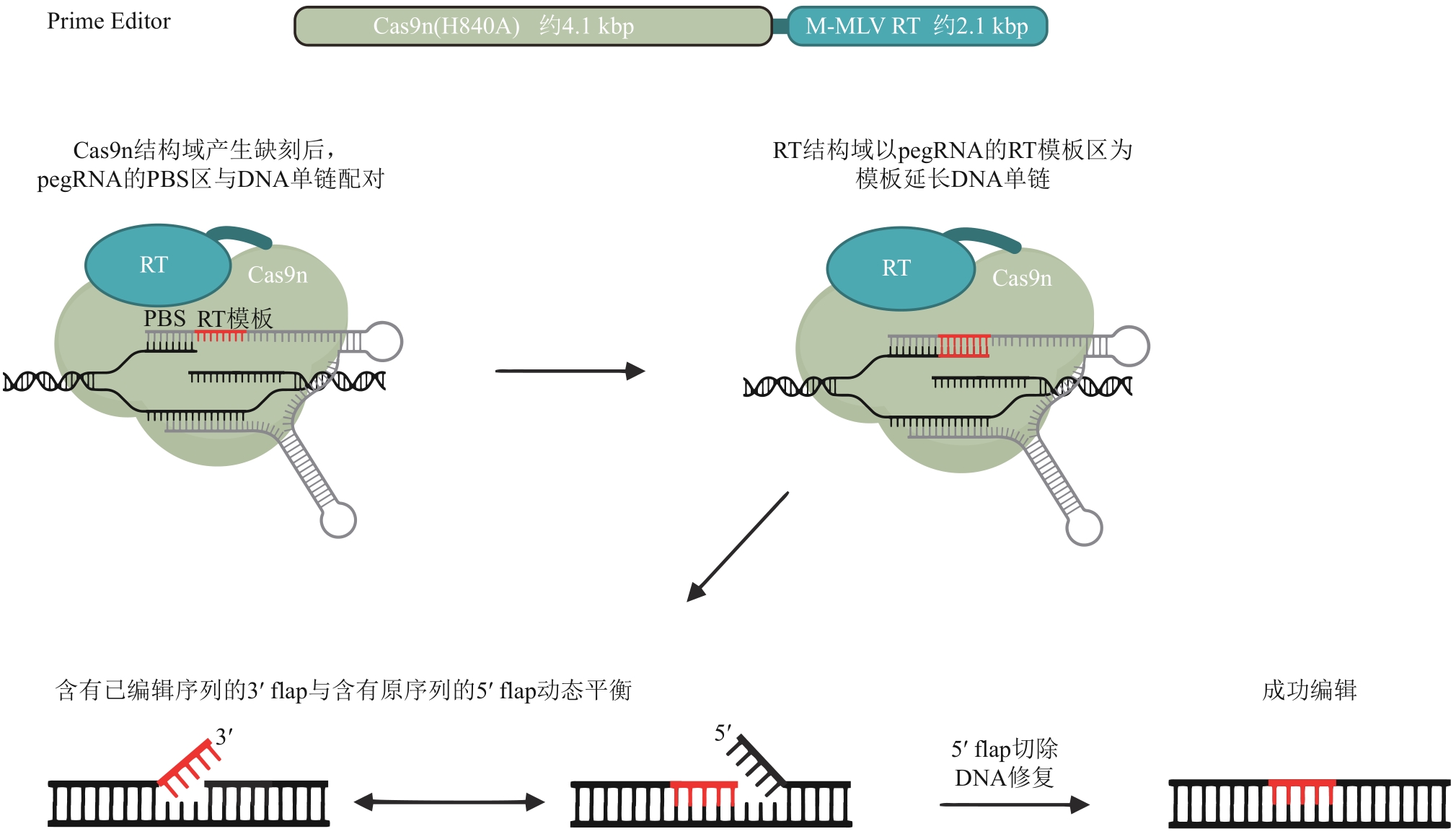
Research progress and biotechnological applications of the prime editing
XU Zhimeng, XIE Zhen
- Department of Automation,Tsinghua University,Beijing National Research Center for Information Science and Technology,Center for Synthesis and Systems Biology,Key Laboratory of Bioinformatics,Ministry of Education,Department of Bioinformatics Research,Beijing 100084,China
-
Received:2023-06-08Revised:2023-09-08Online:2024-03-20Published:2024-02-29 -
Contact:XIE Zhen
引导编辑研究进展及其应用
许志锰, 谢震
- 清华大学自动化系,北京信息科学与技术国家研究中心,生物信息学研究部,生物信息学教育部重点实验室,合成与系统生物学研究中心,北京 100084
-
通讯作者:谢震 -
作者简介:许志锰 (1995—),男,博士研究生。研究方向为合成生物学、基因编辑。 E-mail:xuzm18@mails.tsinghua.edu.cn谢震 (1976—),男,博士,副教授。研究方向为人工分子机器的设计与控制、合成生物学、肿瘤基因组学、肿瘤基因与细胞治疗。 E-mail:zhenxie@tsinghua.edu.cn -
基金资助:国家自然科学基金(32171413)
CLC Number:
Cite this article
XU Zhimeng, XIE Zhen. Research progress and biotechnological applications of the prime editing[J]. Synthetic Biology Journal, 2024, 5(1): 1-15.
许志锰, 谢震. 引导编辑研究进展及其应用[J]. 合成生物学, 2024, 5(1): 1-15.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-038
| 工具名称 | 特点 | 实验细胞系 | 链接 | 参考文献 |
|---|---|---|---|---|
| DeepPE | 给出DeepPE、PE_type、PE_position三个模型的结果 | HEK293FT、HCT116、MDA-MB-231 | http://deepcrispr.info/DeepPE | [ |
| DeepPrime | 将DeepPE扩展到7种细胞系、8种PE | HEK293FT、HCT116、MDA-MB-231、HeLa、DLD1、A549、NIH3T3 | http://deepcrispr.info/DeepPrime/ | [ |
| PRIDICT | 包括ClinVar突变体数据,但输入格式较为复杂 | HEK293T、U2OS、K562 | https://www.pridict.it/ | [ |
| MinsePIE | Spacer、PBS等长度可手动调整,自由度较高 | HEK293T、HAP1 | https://elixir.ut.ee/minsepie/ | [ |
Table 1 Efficiency prediction tools of PE
| 工具名称 | 特点 | 实验细胞系 | 链接 | 参考文献 |
|---|---|---|---|---|
| DeepPE | 给出DeepPE、PE_type、PE_position三个模型的结果 | HEK293FT、HCT116、MDA-MB-231 | http://deepcrispr.info/DeepPE | [ |
| DeepPrime | 将DeepPE扩展到7种细胞系、8种PE | HEK293FT、HCT116、MDA-MB-231、HeLa、DLD1、A549、NIH3T3 | http://deepcrispr.info/DeepPrime/ | [ |
| PRIDICT | 包括ClinVar突变体数据,但输入格式较为复杂 | HEK293T、U2OS、K562 | https://www.pridict.it/ | [ |
| MinsePIE | Spacer、PBS等长度可手动调整,自由度较高 | HEK293T、HAP1 | https://elixir.ut.ee/minsepie/ | [ |
| 名称 | 优化点 | 优化结果 | 实验细胞系或动植物模型 | 参考 文献 |
|---|---|---|---|---|
| PE2* | 优化NLS | 提高编辑效率 | HEK293T | [ |
| PEmax | 更换为SpCas9(R221K/N394K/H840A),优化NLS,优化RT密码子 | 提高编辑效率 | HEK293T、HeLa | [ |
| hyPE2 | 融合Rad51 DNA结合结构域 | 提高编辑效率 | HEK293T、HCT116 | [ |
| CMP-PE-V1 | 融合染色体操纵肽 | 提高编辑效率 | NIH/3T3、C2C12细胞系,C57BL/6N、ICR小鼠 | [ |
| IN-PE2 | 融合短肽 | 提高编辑效率 | HEK293T、U2OS、HCT116 | [ |
| PASTE | 融合Bxb1丝氨酸整合酶 | 可在非分裂细胞基因组中整合超大片段 | HEK293FT、HepG2、K562、原代人外周血CD8 T细胞、原代人肝细胞 | [ |
| ePE | 引入Csy4蛋白 | 提高编辑效率 | HEK293T、HeLa、N2a | [ |
| WT-PE、PEn | Cas9n更换为野生型Cas9 | 增强大片段删除或易位编辑效率,证明了NHEJ途径可用于PE | HEK293T、HeLa、HCT116 | [ |
| 未命名 | 对Cas9n引入N863A或N854A突变 | 减少indel | HEK293T、HeLa、K562 | [ |
| GENEWRITE | 更换LINE-1 RT | 实现大片段插入 | E.coli | [ |
| ePPE | 融合病毒核衣壳蛋白,删除RNaseH结构域 | 提高编辑效率 | Zhonghua11、Kenong199 | [ |
| PE-P3 | 颠倒Cas9n和M-MLV RT | 提高编辑效率 | Nipponbare | [ |
| PE2△Rnh | 删除RNaseH结构域 | 缩小PE尺寸 | HEK293T、Hepa1-6、RAW264.7细胞系,C57BL/6J、FVB/NJ小鼠 | [ |
| sPE | 拆分Cas9n和M-MLV RT | 方便递送和优化 | HEK293T | [ |
PE2-VQR、PE2-VRQR、PE2-VRER、PE2-NG、 PE2-SpG、PE2-SpRY | 更换其他Cas9变体 | 松弛PAM要求,扩展可编辑范围 | HEK293T | [ |
| SaPE2 | SpCas9更换为SaCas9 | 更改PAM要求 | HEK293T、HeLa、K562 | [ |
| CjCas9-PE | SpCas9更换为CjCas9 | 更改PAM要求 | HEK293T | [ |
| FnCas9-PE | SpCas9更换为FnCas9 | 更改PAM要求 | HEK293T、HeLa、K562、U2OS | [ |
| SauriCas9-PE | SpCas9更换为SauriCas9 | 更改PAM要求 | HEK293T | [ |
Table 2 Derivative variants of PE and their characteristics
| 名称 | 优化点 | 优化结果 | 实验细胞系或动植物模型 | 参考 文献 |
|---|---|---|---|---|
| PE2* | 优化NLS | 提高编辑效率 | HEK293T | [ |
| PEmax | 更换为SpCas9(R221K/N394K/H840A),优化NLS,优化RT密码子 | 提高编辑效率 | HEK293T、HeLa | [ |
| hyPE2 | 融合Rad51 DNA结合结构域 | 提高编辑效率 | HEK293T、HCT116 | [ |
| CMP-PE-V1 | 融合染色体操纵肽 | 提高编辑效率 | NIH/3T3、C2C12细胞系,C57BL/6N、ICR小鼠 | [ |
| IN-PE2 | 融合短肽 | 提高编辑效率 | HEK293T、U2OS、HCT116 | [ |
| PASTE | 融合Bxb1丝氨酸整合酶 | 可在非分裂细胞基因组中整合超大片段 | HEK293FT、HepG2、K562、原代人外周血CD8 T细胞、原代人肝细胞 | [ |
| ePE | 引入Csy4蛋白 | 提高编辑效率 | HEK293T、HeLa、N2a | [ |
| WT-PE、PEn | Cas9n更换为野生型Cas9 | 增强大片段删除或易位编辑效率,证明了NHEJ途径可用于PE | HEK293T、HeLa、HCT116 | [ |
| 未命名 | 对Cas9n引入N863A或N854A突变 | 减少indel | HEK293T、HeLa、K562 | [ |
| GENEWRITE | 更换LINE-1 RT | 实现大片段插入 | E.coli | [ |
| ePPE | 融合病毒核衣壳蛋白,删除RNaseH结构域 | 提高编辑效率 | Zhonghua11、Kenong199 | [ |
| PE-P3 | 颠倒Cas9n和M-MLV RT | 提高编辑效率 | Nipponbare | [ |
| PE2△Rnh | 删除RNaseH结构域 | 缩小PE尺寸 | HEK293T、Hepa1-6、RAW264.7细胞系,C57BL/6J、FVB/NJ小鼠 | [ |
| sPE | 拆分Cas9n和M-MLV RT | 方便递送和优化 | HEK293T | [ |
PE2-VQR、PE2-VRQR、PE2-VRER、PE2-NG、 PE2-SpG、PE2-SpRY | 更换其他Cas9变体 | 松弛PAM要求,扩展可编辑范围 | HEK293T | [ |
| SaPE2 | SpCas9更换为SaCas9 | 更改PAM要求 | HEK293T、HeLa、K562 | [ |
| CjCas9-PE | SpCas9更换为CjCas9 | 更改PAM要求 | HEK293T | [ |
| FnCas9-PE | SpCas9更换为FnCas9 | 更改PAM要求 | HEK293T、HeLa、K562、U2OS | [ |
| SauriCas9-PE | SpCas9更换为SauriCas9 | 更改PAM要求 | HEK293T | [ |
| 名称 | pegRNA优化方式 | 参考 文献 |
|---|---|---|
| epegRNA | 3′端添加tevopreQ1或mpknot基序以防降解 | [ |
| xrPE | 3′端添加xrRNA继续以防降解 | [ |
| G-PE | 3′端添加G-quadruplex基序以防降解 | [ |
| ePE | 3′端添加Csy4基序以防环化 | [ |
| apegRNA | 优化hairpin | [ |
| spegRNA | RT模板引入统一突变以改善二级结构 | [ |
Table 3 Optimization methods for pegRNA
| 名称 | pegRNA优化方式 | 参考 文献 |
|---|---|---|
| epegRNA | 3′端添加tevopreQ1或mpknot基序以防降解 | [ |
| xrPE | 3′端添加xrRNA继续以防降解 | [ |
| G-PE | 3′端添加G-quadruplex基序以防降解 | [ |
| ePE | 3′端添加Csy4基序以防环化 | [ |
| apegRNA | 优化hairpin | [ |
| spegRNA | RT模板引入统一突变以改善二级结构 | [ |
| 名称 | 特点 | 参考文献 |
|---|---|---|
| 无 | 在水稻中提高碱基替换、小片段插入删除编辑效率2.9~17.4倍 | [ |
| HOPE | 提高碱基替换、小片段插入删除编辑效率和精确度 | [ |
| twinPE | 大片段插入、删除、替换,可与Bxb1丝氨酸重组酶联用 | [ |
| PRIME-Del | 高达10 kb的大片段删除,并允许再删除同时插入短片段 | [ |
| GRAND | 高达1 kb的大片段插入 | [ |
| PEDAR | 高达1~10 kb的大片段删除,并允许替换为高达60 bp的片段 | [ |
| PETI | 染色体易位、颠倒 | [ |
| WT-PE | 野生型Cas9引入DSB,实现大片段删除和易位 | [ |
| Bi-PE | 大片段删除 | [ |
Table 4 Paired pegRNA
| 名称 | 特点 | 参考文献 |
|---|---|---|
| 无 | 在水稻中提高碱基替换、小片段插入删除编辑效率2.9~17.4倍 | [ |
| HOPE | 提高碱基替换、小片段插入删除编辑效率和精确度 | [ |
| twinPE | 大片段插入、删除、替换,可与Bxb1丝氨酸重组酶联用 | [ |
| PRIME-Del | 高达10 kb的大片段删除,并允许再删除同时插入短片段 | [ |
| GRAND | 高达1 kb的大片段插入 | [ |
| PEDAR | 高达1~10 kb的大片段删除,并允许替换为高达60 bp的片段 | [ |
| PETI | 染色体易位、颠倒 | [ |
| WT-PE | 野生型Cas9引入DSB,实现大片段删除和易位 | [ |
| Bi-PE | 大片段删除 | [ |
| 名称 | 软件特点 | 适用范围 | 参考文献 |
|---|---|---|---|
| PINE-CONE | 仅有离线可执行文件,可部署在本地,支持Win和Mac | 仅支持PE | [ |
| PnB Designer | 在线,PBS、RTT长度需自定 | 支持多种基因组,兼容BE | [ |
| Easy-Prime | 在线,输入格式有要求 | 仅支持PE | [ |
| Prime Editor Designer | 在线,无自定义序列设计 | 仅对NCBI、ClinVar数据库基因设计,支持两种Cas9变体 | [ |
| pegFinder | 在线,输入序列格式直观 | 支持对自定义序列设计,支持三种Cas9变体 | [ |
| Plant Peg Designer | 在线,输入序列格式直观 | 针对植物,支持两种Cas9变体和双pegRNA策略,支持对自定义序列设计 | [ |
| PrimeDesign | 在线,输入序列格式直观,支持预览二级结构 | 支持对自定义序列设计 | [ |
| PETAL | 在线,输入序列格式直观 | 需与其PEA1单质粒系统配合使用 | [ |
| pegIT | 在线,输出分子克隆序列参考 | 支持多种基因组和ClinVar,支持五种Cas9 | [ |
| PE-designer | 在线,输入序列格式直观,支持E-mail收取结果 | 支持极丰富的Cas9变体和基因组 | [ |
Table 5 Assisted design tools for pegRNA
| 名称 | 软件特点 | 适用范围 | 参考文献 |
|---|---|---|---|
| PINE-CONE | 仅有离线可执行文件,可部署在本地,支持Win和Mac | 仅支持PE | [ |
| PnB Designer | 在线,PBS、RTT长度需自定 | 支持多种基因组,兼容BE | [ |
| Easy-Prime | 在线,输入格式有要求 | 仅支持PE | [ |
| Prime Editor Designer | 在线,无自定义序列设计 | 仅对NCBI、ClinVar数据库基因设计,支持两种Cas9变体 | [ |
| pegFinder | 在线,输入序列格式直观 | 支持对自定义序列设计,支持三种Cas9变体 | [ |
| Plant Peg Designer | 在线,输入序列格式直观 | 针对植物,支持两种Cas9变体和双pegRNA策略,支持对自定义序列设计 | [ |
| PrimeDesign | 在线,输入序列格式直观,支持预览二级结构 | 支持对自定义序列设计 | [ |
| PETAL | 在线,输入序列格式直观 | 需与其PEA1单质粒系统配合使用 | [ |
| pegIT | 在线,输出分子克隆序列参考 | 支持多种基因组和ClinVar,支持五种Cas9 | [ |
| PE-designer | 在线,输入序列格式直观,支持E-mail收取结果 | 支持极丰富的Cas9变体和基因组 | [ |
| 1 | ANZALONE A V, KOBLAN L W, LIU D R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nature Biotechnology, 2020, 38(7): 824-844. |
| 2 | KOMOR A C, BADRAN A H, LIU D R. CRISPR-based technologies for the manipulation of eukaryotic genomes[J]. Cell, 2017, 168(1/2): 20-36. |
| 3 | WILLIAMS A B, SCHUMACHER B. p53 in the DNA-damage-repair process[J]. Cold Spring Harbor Perspectives in Medicine, 2016, 6(5): a026070. |
| 4 | HAAPANIEMI E, BOTLA S, PERSSON J, et al. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response[J]. Nature Medicine, 2018, 24(7): 927-930. |
| 5 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| 6 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| 7 | KURT I C, ZHOU R H, IYER S, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells[J]. Nature Biotechnology, 2021, 39(1): 41-46. |
| 8 | CHEN L, HONG M J, LUAN C M, et al. Adenine transversion editors enable precise, efficient A•T-to-C•G base editing in mammalian cells and embryos[J/OL]. Nature Biotechnology, 2023[2023-07-14]. . |
| 9 | TONG H W, LIU N N, WEI Y H, et al. Programmable deaminase-free base editors for G-to-Y conversion by engineered glycosylase[J]. National Science Review, 2023, 10(8): nwad143. |
| 10 | HAN D Y, XIAO Q Q, WANG Y F, et al. Development of miniature base editors using engineered IscB nickase[J]. Nature Methods, 2023, 20(7): 1029-1036. |
| 11 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| 12 | KIM H K, YU G, PARK J, et al. Predicting the efficiency of prime editing guide RNAs in human cells[J]. Nature Biotechnology, 2021, 39(2): 198-206. |
| 13 | LIN Q P, ZONG Y, XUE C X, et al. Prime genome editing in rice and wheat[J]. Nature Biotechnology, 2020, 38(5): 582-585. |
| 14 | LIN Q P, JIN S, ZONG Y, et al. High-efficiency prime editing with optimized, paired pegRNAs in plants[J]. Nature Biotechnology, 2021, 39(8): 923-927. |
| 15 | PONNIENSELVAN K, LIU P P, NYALILE T, et al. Reducing the inherent auto-inhibitory interaction within the pegRNA enhances prime editing efficiency[J]. Nucleic Acids Research, 2023, 51(13): 6966-6980. |
| 16 | KIM H K, LEE S, KIM Y, et al. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells[J]. Nature Biomedical Engineering, 2020, 4(1): 111-124. |
| 17 | CHARI R, MALI P, MOOSBURNER M, et al. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach[J]. Nature Methods, 2015, 12(9): 823-826. |
| 18 | KIM H K, SONG M, LEE J N, et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity[J]. Nature Methods, 2017, 14(2): 153-159. |
| 19 | KIM H K, MIN S, SONG M, et al. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity[J]. Nature Biotechnology, 2018, 36(3): 239-241. |
| 20 | KIM H K, KIM Y, LEE S, et al. SpCas9 activity prediction by DeepSpCas9, a deep learning-based model with high generalization performance[J]. Science Advances, 2019, 5(11): eaax9249. |
| 21 | YU G, KIM H K, PARK J, et al. Prediction of efficiencies for diverse prime editing systems in multiple cell types[J]. Cell, 2023, 186(10): 2256-2272.e23. |
| 22 | MATHIS N, ALLAM A, KISSLING L, et al. Predicting prime editing efficiency and product purity by deep learning[J]. Nature Biotechnology, 2023, 41(8): 1151-1159. |
| 23 | KOEPPEL J, WELLER J, PEETS E M, et al. Prediction of prime editing insertion efficiencies using sequence features and DNA repair determinants[J]. Nature Biotechnology, 2023, 41: 1446-1456. |
| 24 | KIM D Y, MOON S B, KO J H, et al. Unbiased investigation of specificities of prime editing systems in human cells[J]. Nucleic Acids Research, 2020, 48(18): 10576-10589. |
| 25 | HABIB O, HABIB G, HWANG G H, et al. Comprehensive analysis of prime editing outcomes in human embryonic stem cells[J]. Nucleic Acids Research, 2022, 50(2): 1187-1197. |
| 26 | GAO R Z, FU Z C, LI X Y, et al. Genomic and transcriptomic analyses of prime editing guide RNA-independent off-target effects by prime editors[J]. The CRISPR Journal, 2022, 5(2): 276-293. |
| 27 | KWON J, KIM M, BAE S, et al. TAPE-seq is a cell-based method for predicting genome-wide off-target effects of prime editor[J]. Nature Communications, 2022, 13: 7975. |
| 28 | LIANG S Q, LIU P P, PONNIENSELVAN K, et al. Genome-wide profiling of prime editor off-target sites in vitro and in vivo using PE-tag[J]. Nature Methods, 2023, 20(6): 898-907. |
| 29 | JIN S, LIN Q P, LUO Y F, et al. Genome-wide specificity of prime editors in plants[J]. Nature Biotechnology, 2021, 39(10): 1292-1299. |
| 30 | LIU P P, LIANG S Q, ZHENG C W, et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice[J]. Nature Communications, 2021, 12: 2121. |
| 31 | CHEN P J, HUSSMANN J A, YAN J, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes[J]. Cell, 2021, 184(22): 5635-5652.e29. |
| 32 | SONG M, LIM J M, MIN S, et al. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain[J]. Nature Communications, 2021, 12: 5617. |
| 33 | PARK S J, JEONG T Y, SHIN S K, et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor[J]. Genome Biology, 2021, 22(1): 170. |
| 34 | VELIMIROVIC M, ZANETTI L C, SHEN M W, et al. Peptide fusion improves prime editing efficiency[J]. Nature Communications, 2022, 13(1): 3512. |
| 35 | YARNALL M T N, IOANNIDI E I, SCHMITT-ULMS C, et al. Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases[J]. Nature Biotechnology, 2023, 41(4): 500-512. |
| 36 | LIU Y, YANG G, HUANG S H, et al. Enhancing prime editing by Csy4-mediated processing of pegRNA[J]. Cell Research, 2021, 31(10): 1134-1136. |
| 37 | TAO R, WANG Y H, HU Y, et al. WT-PE: prime editing with nuclease wild-type Cas9 enables versatile large-scale genome editing[J]. Signal Transduction and Targeted Therapy, 2022, 7: 108. |
| 38 | PETERKA M, AKRAP N, LI S Y, et al. Harnessing DSB repair to promote efficient homology-dependent and-independent prime editing[J]. Nature Communications, 2022, 13(1): 1240. |
| 39 | LEE J, LIM K, KIM A, et al. Prime editing with genuine Cas9 nickases minimizes unwanted indels[J]. Nature Communications, 2023, 14: 1786. |
| 40 | MANOJ F, TAI L W, WANG K S M, et al. Targeted insertion of large genetic payloads using Cas directed LINE-1 reverse transcriptase[J]. Scientific Reports, 2021, 11: 23625. |
| 41 | ZONG Y, LIU Y J, XUE C X, et al. An engineered prime editor with enhanced editing efficiency in plants[J]. Nature Biotechnology, 2022, 40(9): 1394-1402. |
| 42 | XU W, YANG Y X, YANG B Y, et al. A design optimized prime editor with expanded scope and capability in plants[J]. Nature Plants, 2022, 8(1): 45-52. |
| 43 | ZHENG C W, LIANG S Q, LIU B, et al. A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver[J]. Molecular Therapy, 2022, 30(3): 1343-1351. |
| 44 | BÖCK D, ROTHGANGL T, VILLIGER L, et al. In vivo prime editing of a metabolic liver disease in mice[J]. Science Translational Medicine, 2022, 14(636): eabl9238. |
| 45 | GAO Z L, RAVENDRAN S, MIKKELSEN N S, et al. A truncated reverse transcriptase enhances prime editing by split AAV vectors[J]. Molecular Therapy, 2022, 30(9): 2942-2951. |
| 46 | LIU B, DONG X L, CHENG H Y, et al. A split prime editor with untethered reverse transcriptase and circular RNA template[J]. Nature Biotechnology, 2022, 40(9): 1388-1393. |
| 47 | KWEON J, YOON J K, JANG A H, et al. Engineered prime editors with PAM flexibility[J]. Molecular Therapy, 2021, 29(6): 2001-2007. |
| 48 | OH Y, LEE W J, HUR J K, et al. Expansion of the prime editing modality with Cas9 from Francisella novicida [J]. Genome Biology, 2022, 23(1): 92. |
| 49 | ZHANG X H, CHEN L, ZHU B Y, et al. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain[J]. Nature Cell Biology, 2020, 22(6): 740-750. |
| 50 | CHEN R H, CAO Y, LIU Y J, et al. Enhancement of a prime editing system via optimal recruitment of the pioneer transcription factor P65[J]. Nature Communications, 2023, 14: 257. |
| 51 | VELIMIROVIC M, ZANETTI L C, SHEN M W, et al. Peptide fusion improves prime editing efficiency[J]. Nature Communications, 2022, 13(1): 3512. |
| 52 | GRÜNEWALD J, MILLER B R, SZALAY R N, et al. Engineered CRISPR prime editors with compact, untethered reverse transcriptases[J]. Nature Biotechnology, 2023, 41(3): 337-343. |
| 53 | NELSON J W, RANDOLPH P B, SHEN S P, et al. Engineered pegRNAs improve prime editing efficiency[J]. Nature Biotechnology, 2022, 40(3): 402-410. |
| 54 | ZHANG G Q, LIU Y, HUANG S S, et al. Enhancement of prime editing via xrRNA motif-joined pegRNA[J]. Nature Communications, 2022, 13: 1856. |
| 55 | LI X Y, WANG X, SUN W J, et al. Enhancing prime editing efficiency by modified pegRNA with RNA G-quadruplexes[J]. Journal of Molecular Cell Biology, 2022, 14(4): mjac022. |
| 56 | LI X S, ZHOU L N, GAO B Q, et al. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure[J]. Nature Communications, 2022, 13: 1669. |
| 57 | RIESENBERG S, HELMBRECHT N, KANIS P, et al. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage[J]. Nature Communications, 2022, 13: 489. |
| 58 | ZHUANG Y, LIU J L, WU H, et al. Increasing the efficiency and precision of prime editing with guide RNA pairs[J]. Nature Chemical Biology, 2022, 18(1): 29-37. |
| 59 | ANZALONE A V, GAO X D, PODRACKY C J, et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing[J]. Nature Biotechnology, 2022, 40(5): 731-740. |
| 60 | CHOI J, CHEN W, SUITER C C, et al. Precise genomic deletions using paired prime editing[J]. Nature Biotechnology, 2022, 40(2): 218-226. |
| 61 | WANG J L, HE Z, WANG G Q, et al. Efficient targeted insertion of large DNA fragments without DNA donors[J]. Nature Methods, 2022, 19(3): 331-340. |
| 62 | JIANG T T, ZHANG X O, WENG Z P, et al. Deletion and replacement of long genomic sequences using prime editing[J]. Nature Biotechnology, 2022, 40(2): 227-234. |
| 63 | KWEON J, HWANG H Y, RYU H, et al. Targeted genomic translocations and inversions generated using a paired prime editing strategy[J]. Molecular Therapy, 2023, 31(1): 249-259. |
| 64 | TAO R, WANG Y H, JIAO Y G, et al. Bi-PE: bi-directional priming improves CRISPR/Cas9 prime editing in mammalian cells[J]. Nucleic Acids Research, 2022, 50(11): 6423-6434. |
| 65 | FERREIRA DA SILVA J, OLIVEIRA G P, ARASA-VERGE E A, et al. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair[J]. Nature Communications, 2022, 13: 760. |
| 66 | LIU N, ZHOU L F, LIN G F, et al. HDAC inhibitors improve CRISPR-Cas9 mediated prime editing and base editing[J]. Molecular Therapy - Nucleic Acids, 2022, 30: 173. |
| 67 | IHRY R J, WORRINGER K A, SALICK M R, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells[J]. Nature Medicine, 2018, 24(7): 939-946. |
| 68 | LI M, ZHONG A, WU Y J, et al. Transient inhibition of p53 enhances prime editing and cytosine base-editing efficiencies in human pluripotent stem cells[J]. Nature Communications, 2022, 13: 6354. |
| 69 | HUANG S S, ZHANG Z W, TAO W Y, et al. Broadening prime editing toolkits using RNA-Pol-Ⅱ-driven engineered pegRNA[J]. Molecular Therapy, 2022, 30(9): 2923-2932. |
| 70 | STANDAGE-BEIER K, TEKEL S J, BRAFMAN D A, et al. Prime editing guide RNA design automation using PINE-CONE[J]. ACS Synthetic Biology, 2021, 10(2): 422-427. |
| 71 | SIEGNER S M, KARASU M E, SCHRÖDER M S, et al. PnB Designer: a web application to design prime and base editor guide RNAs for animals and plants[J]. BMC Bioinformatics, 2021, 22(1): 101. |
| 72 | LI Y C, CHEN J J, TSAI S Q, et al. Easy-Prime: a machine learning-based prime editor design tool[J]. Genome Biology, 2021, 22(1): 235. |
| 73 | MORRIS J A, RAHMAN J A, GUO X Y, et al. Automated design of CRISPR prime editors for 56, 000 human pathogenic variants[J]. iScience, 2021, 24(11): 103380. |
| 74 | CHOW R D, CHEN J S, SHEN J, et al. A web tool for the design of prime-editing guide RNAs[J]. Nature Biomedical Engineering, 2021, 5(2): 190-194. |
| 75 | HSU J Y, GRÜNEWALD J, SZALAY R, et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs[J]. Nature Communications, 2021, 12: 1034. |
| 76 | ADIKUSUMA F, LUSHINGTON C, ARUDKUMAR J, et al. Optimized nickase- and nuclease-based prime editing in human and mouse cells[J]. Nucleic Acids Research, 2021, 49(18): 10785-10795. |
| 77 | ANDERSON M V, HALDRUP J, THOMSEN E A, et al. pegIT-a web-based design tool for prime editing[J]. Nucleic Acids Research, 2021, 49(W1): W505-W509. |
| 78 | HWANG G H, JEONG Y K, HABIB O, et al. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing[J]. Nucleic Acids Research, 2021, 49(W1): W499-W504. |
| 79 | LI H Q, BUSQUETS O, VERMA Y, et al. Highly efficient generation of isogenic pluripotent stem cell models using prime editing[J]. eLife, 2022, 11: 79208. |
| 80 | SÜRÜN D, SCHNEIDER A, MIRCETIC J, et al. Efficient generation and correction of mutations in human iPS cells utilizing mRNAs of CRISPR base editors and prime editors[J]. Genes, 2020, 11(5): 511. |
| 81 | PETRI K, ZHANG W T, MA J Y, et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells[J]. Nature Biotechnology, 2022, 40(2): 189-193. |
| 82 | WOLFF J H, HALDRUP J, THOMSEN E A, et al. piggyPrime: high-efficacy prime editing in human cells using piggyBac-based DNA transposition[J]. Frontiers in Genome Editing, 2021, 3: 786893. |
| 83 | EGGENSCHWILER R, GSCHWENDTBERGER T, FELSKI C, et al. A selectable all-in-one CRISPR prime editing piggyBac transposon allows for highly efficient gene editing in human cell lines[J]. Scientific Reports, 2021, 11: 22154. |
| 84 | ZHI S Y, CHEN Y X, WU G L, et al. Dual-AAV delivering split prime editor system for in vivo genome editing[J]. Molecular Therapy, 2022, 30(1): 283-294. |
| 85 | SUN L W, LI J, XIAO X. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization[J]. Nature Medicine, 2000, 6(5): 599-602. |
| 86 | JANG H, JO D H, CHO C S, et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases[J]. Nature Biomedical Engineering, 2022, 6(2): 181-194. |
| 87 | DAVIS J R, BANSKOTA S, LEVY J M, et al. Efficient prime editing in mouse brain, liver and heart with dual AAVs[J/OL]. Nature Biotechnology, 2023[2023-07-10]. . |
| 88 | WANG Q, LIU J, JANSSEN J M, et al. Broadening the reach and investigating the potential of prime editors through fully viral gene-deleted adenoviral vector delivery[J]. Nucleic Acids Research, 2021, 49(20): 11986-12001. |
| 89 | AULICINO F, PELOSSE M, TOELZER C, et al. Highly efficient CRISPR-mediated large DNA docking and multiplexed prime editing using a single baculovirus[J]. Nucleic Acids Research, 2022, 50(13): 7783-7799. |
| 90 | LIU Y, LI X Y, HE S T, et al. Efficient generation of mouse models with the prime editing system[J]. Cell Discovery, 2020, 6: 27. |
| 91 | GAO P, LYU Q, GHANAM A R, et al. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression[J]. Genome Biology, 2021, 22(1): 83. |
| 92 | LIN J X, LIU X C, LU Z Y, et al. Modeling a cataract disorder in mice with prime editing[J]. Molecular Therapy Nucleic Acids, 2021, 25: 494-501. |
| 93 | QIAN Y Q, ZHAO D, SUI T T, et al. Efficient and precise generation of Tay-Sachs disease model in rabbit by prime editing system[J]. Cell Discovery, 2021, 7: 50. |
| 94 | KIM D E, LEE J H, JI K BIN, et al. Prime editor-mediated correction of a pathogenic mutation in purebred dogs[J]. Scientific Reports, 2022, 12: 12905. |
| 95 | ATSUTA Y, SUZUKI K, IIKAWA H, et al. Prime editing in chicken fibroblasts and primordial germ cells[J]. Development, Growth & Differentiation, 2022, 64(9): 548-557. |
| 96 | BOSCH J A, BIRCHAK G, PERRIMON N. Precise genome engineering in Drosophila using prime editing[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(1): e2021996118. |
| 97 | KIM Y, HONG S A, YU J, et al. Adenine base editing and prime editing of chemically derived hepatic progenitors rescue genetic liver disease[J]. Cell Stem Cell, 2021, 28(9): 1614-1624.e5. |
| 98 | CHEMELLO F, CHAI A C, LI H, et al. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing[J]. Science Advances, 2021, 7(18): eabg4910. |
| 99 | HAPPI MBAKAM C, ROUSSEAU J, LU Y Y, et al. Prime editing optimized RTT permits the correction of the c. 8713C>T mutation in DMD gene[J]. Molecular Therapy Nucleic Acids, 2022, 30: 272-285. |
| 100 | LV X J, ZHENG Z, ZHI X, et al. Identification of RPGR ORF15 mutation for X-linked retinitis pigmentosa in a large Chinese family and in vitro correction with prime editor[J]. Gene Therapy, 2023, 30(1/2): 160-166. |
| 101 | HONG S A, KIM S E, LEE A Y, et al. Therapeutic base editing and prime editing of COL7A1 mutations in recessive dystrophic epidermolysis bullosa[J]. Molecular Therapy, 2022, 30(8): 2664-2679. |
| 102 | ZHOU M J, TANG S Q, DUAN N N, et al. Targeted-deletion of a tiny sequence via prime editing to restore SMN expression[J]. International Journal of Molecular Sciences, 2022, 23(14): 7941. |
| 103 | ZHANG H K, ZHOU Q, CHEN H Y, et al. Prime editor 3 mediated beta-thalassemia mutations of the HBB gene in human erythroid progenitor cells[J]. International Journal of Molecular Sciences, 2022, 23(9): 5002. |
| 104 | ZHANG H K, SUN R L, FEI J, et al. Correction of beta-thalassemia IVS-Ⅱ-654 mutation in a mouse model using prime editing[J]. International Journal of Molecular Sciences, 2022, 23(11): 5948. |
| 105 | ABUHAMAD A Y, MOHAMAD ZAMBERI N N, LING S E, et al. Reverting TP53 mutation in breast cancer cells: prime editing workflow and technical considerations[J]. Cells, 2022, 11(10): 1612. |
| 106 | TREMBLAY G, ROUSSEAU J, MBAKAM C H, et al. Insertion of the Icelandic mutation (A673T) by prime editing: a potential preventive treatment for familial and sporadic Alzheimer's disease[J]. The CRISPR Journal, 2022, 5(1): 109-122. |
| 107 | XU R F, LI J, LIU X S, et al. Development of plant prime-editing systems for precise genome editing[J]. Plant Communications, 2020, 1(3): 100043. |
| 108 | LI Z S, MA R, LIU D, et al. A straightforward plant prime editing system enabled highly efficient precise editing of rice Waxy gene[J]. Plant Science, 2022, 323: 111400. |
| 109 | JIN S, LIN Q P, GAO Q, et al. Optimized prime editing in monocot plants using PlantPegDesigner and engineered plant prime editors (ePPEs)[J]. Nature Protocols, 2023, 18(3): 831-853. |
| 110 | XU R F, LIU X S, LI J, et al. Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice[J]. Nature Plants, 2021, 7(7): 888-892. |
| 111 | JIANG Y Y, CHAI Y P, LU M H, et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize[J]. Genome Biology, 2020, 21(1): 257. |
| 112 | QIAO D X, WANG J Y, LU M H, et al. Optimized prime editing efficiently generates heritable mutations in maize[J]. Journal of Integrative Plant Biology, 2023, 65(4): 900-906. |
| 113 | JIANG Z Z, ABDULLAH, ZHANG S, et al. Development and optimization of CRISPR prime editing system in photoautotrophic cells[J]. Molecules, 2022, 27(6): 1758. |
| 114 | LU Y M, TIAN Y F, SHEN R D, et al. Precise genome modification in tomato using an improved prime editing system[J]. Plant Biotechnology Journal, 2021, 19(3): 415-417. |
| 115 | PERROUD P F, GUYON-DEBAST A, VEILLET F, et al. Prime Editing in the model plant Physcomitrium patens and its potential in the tetraploid potato[J]. Plant Science, 2022, 316: 111162. |
| 116 | BISWAS S, BRIDGELAND A, IRUM S, et al. Optimization of prime editing in rice, peanut, chickpea, and cowpea protoplasts by restoration of GFP activity[J]. International Journal of Molecular Sciences, 2022, 23(17): 9809. |
| [1] | CHEN Yingying, LIU Yang, SHI Junjie, MA Junying, JU Jianhua. CRISPR/Cas systems and their applications in gene editing with filamentous fungi [J]. Synthetic Biology Journal, 2024, 5(3): 672-693. |
| [2] | Yaru CHEN, Yingxiu CAO, Hao SONG. Advances and applications of gene editing and transcriptional regulation in electroactive microorganisms [J]. Synthetic Biology Journal, 2023, 4(6): 1281-1299. |
| [3] | Jicong LIN, Gen ZOU, Hongmin LIU, Yongjun WEI. Application of CRISPR/Cas genome editing technology in the synthesis of secondary metabolites of filamentous fungi [J]. Synthetic Biology Journal, 2023, 4(4): 738-755. |
| [4] | Yuanhuan YU, Yang ZHOU, Xinyi WANG, Deqiang KONG, Haifeng YE. Advances in optogenetics for biomedical research [J]. Synthetic Biology Journal, 2023, 4(1): 102-140. |
| [5] | Ke LIU, Guihong LIN, Kun LIU, Wei ZHOU, Fengqing WANG, Dongzhi WEI. Mining, engineering and functional expansion of CRISPR/Cas systems [J]. Synthetic Biology Journal, 2023, 4(1): 47-66. |
| [6] | Liya LIANG, Rongming LIU. Protein engineering of DNA targeting type Ⅱ CRISPR/Cas systems [J]. Synthetic Biology Journal, 2023, 4(1): 86-101. |
| [7] | Jiaxin LIU, Chi CHENG, Xinqi LI, Chaojun WANG, Ying ZHANG, Chuang XUE. Recent progress in the molecular genetic modification tools of Clostridium [J]. Synthetic Biology Journal, 2022, 3(6): 1201-1217. |
| [8] | Jiacheng BI, Zhigang TIAN. Synthetic immunology and future NK cell immunotherapy [J]. Synthetic Biology Journal, 2022, 3(1): 22-34. |
| [9] | Han XIAO, Yixin LIU. Progress and challenge of the CRISPR-Cas system in gene editing for filamentous fungi [J]. Synthetic Biology Journal, 2021, 2(2): 274-286. |
| [10] | Yang LI, Xiaolin SHEN, Xinxiao SUN, Qipeng YUAN, Yajun YAN, Jia WANG. Advances of CRISPR gene editing in microbial synthetic biology [J]. Synthetic Biology Journal, 2021, 2(1): 106-120. |
| [11] | Yongfu YANG, Binan GENG, Haoyue SONG, Qiaoning HE, Mingxiong HE, Jie BAO, Fengwu BAI, Shihui YANG. Progress and perspectives on developing Zymomonas mobilis as a chassis cell [J]. Synthetic Biology Journal, 2021, 2(1): 59-90. |
| [12] | Zhongzheng CAO, Xinyi ZHANG, Yiyuan XU, Zhuo ZHOU, Wensheng WEI. Genome editing technology and its applications in synthetic biology [J]. Synthetic Biology Journal, 2020, 1(4): 413-426. |
| [13] | Lu LIN, Xueqin LV, Yanfeng LIU, Guocheng DU, Jian CHEN, Long LIU. Advances in design, construction and applications of Bacillus subtilis chassis cells [J]. Synthetic Biology Journal, 2020, 1(2): 247-265. |
| [14] | Bo ZHANG, Yongshuo MA, Yi SHANG, Sanwen HUANG. Recent advances in plant synthetic biology [J]. Synthetic Biology Journal, 2020, 1(2): 121-140. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||