Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (2): 247-265.DOI: 10.12211/2096-8280.2020-030
• Invited Review • Previous Articles
Advances in design, construction and applications of Bacillus subtilis chassis cells
LIN Lu1, LV Xueqin1, LIU Yanfeng1, DU Guocheng1, CHEN Jian2, LIU Long1
- 1.Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, Jiangnan University, Wuxi 214122, Jiangsu, China
2.Key Laboratory of Industrial Biotechnology, Ministry of Education, Jiangnan University, Wuxi 214122, Jiangsu, China
-
Received:2020-03-13Revised:2020-04-08Online:2020-08-04Published:2020-04-30
枯草芽孢杆菌底盘细胞的设计、构建与应用
林璐1, 吕雪芹1, 刘延峰1, 堵国成1, 陈坚2, 刘龙1
- 1.江南大学糖化学与生物技术教育部重点实验室 江苏 无锡 214122
2.江南大学工业生物技术教育部重点实验室 江苏 无锡 214122
-
作者简介:林璐(1993—),女,博士研究生,研究方向为微生物代谢工程。E-mail:linlu@stu.jiangnan.edu.cn
刘龙(1980—),男,博士,教授,研究方向为代谢工程与合成生物学。E-mail:longliu@jiangnan.edu.cn -
基金资助:国家自然科学基金优秀青年基金(31622001);国家重点研发计划(2018YFA0900300)
CLC Number:
Cite this article
LIN Lu, LV Xueqin, LIU Yanfeng, DU Guocheng, CHEN Jian, LIU Long. Advances in design, construction and applications of Bacillus subtilis chassis cells[J]. Synthetic Biology Journal, 2020, 1(2): 247-265.
林璐, 吕雪芹, 刘延峰, 堵国成, 陈坚, 刘龙. 枯草芽孢杆菌底盘细胞的设计、构建与应用[J]. 合成生物学, 2020, 1(2): 247-265.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-030
| 类别 | 构建方法 | 功能特性 | 文献 |
|---|---|---|---|
| 基因编辑 | 基因组简化 | 利用无痕基因敲除技术构建多种基因组简化菌株 | [ |
| CRISPR/Cas9迭代系统 | 实现基因敲除、点突变和基因敲入,避免脱靶效应 | [ | |
| CRISPR-Cas9工具包 | 实现基因敲除、多基因敲入和转录抑制 | [ | |
| CAMERS-B系统 | 通过设计crRNA阵列实现多基因编辑和调控 | [ | |
| 内源性启动子库 | 基于细胞生长构建的内源性启动子文库 | [ | |
| 模块化操作单元 | 在转录翻译和蛋白水解水平上共同调节基因和蛋白的表达 | [ | |
| DNA支架 | 通过DNA支架调节途径酶的空间比例和方向 | [ | |
| RNA支架 | 通过RNA支架与特异性蛋白结合,转录抑制靶基因的表达 | [ | |
| 内源性空间支架 | 通过FMMs空间支架构建FMMs-多酶复合物系统 | [ | |
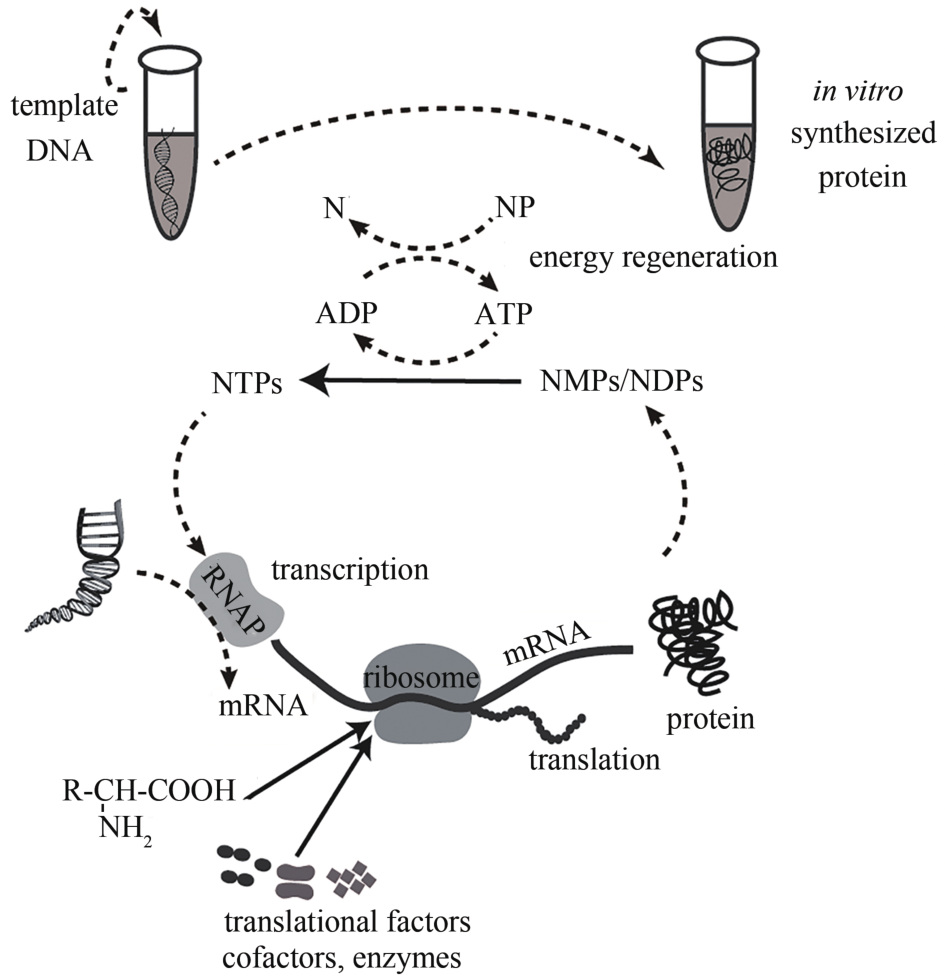
| 无细胞蛋白表达系统 | 以外源DNA为模板,利用细胞裂解物,可在体外表达蛋白质 | [ | |
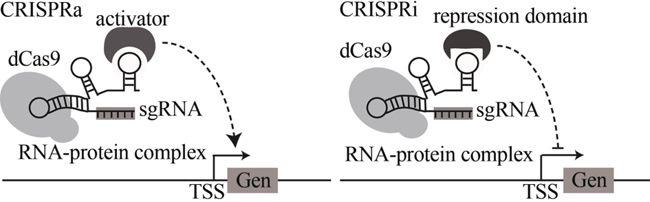
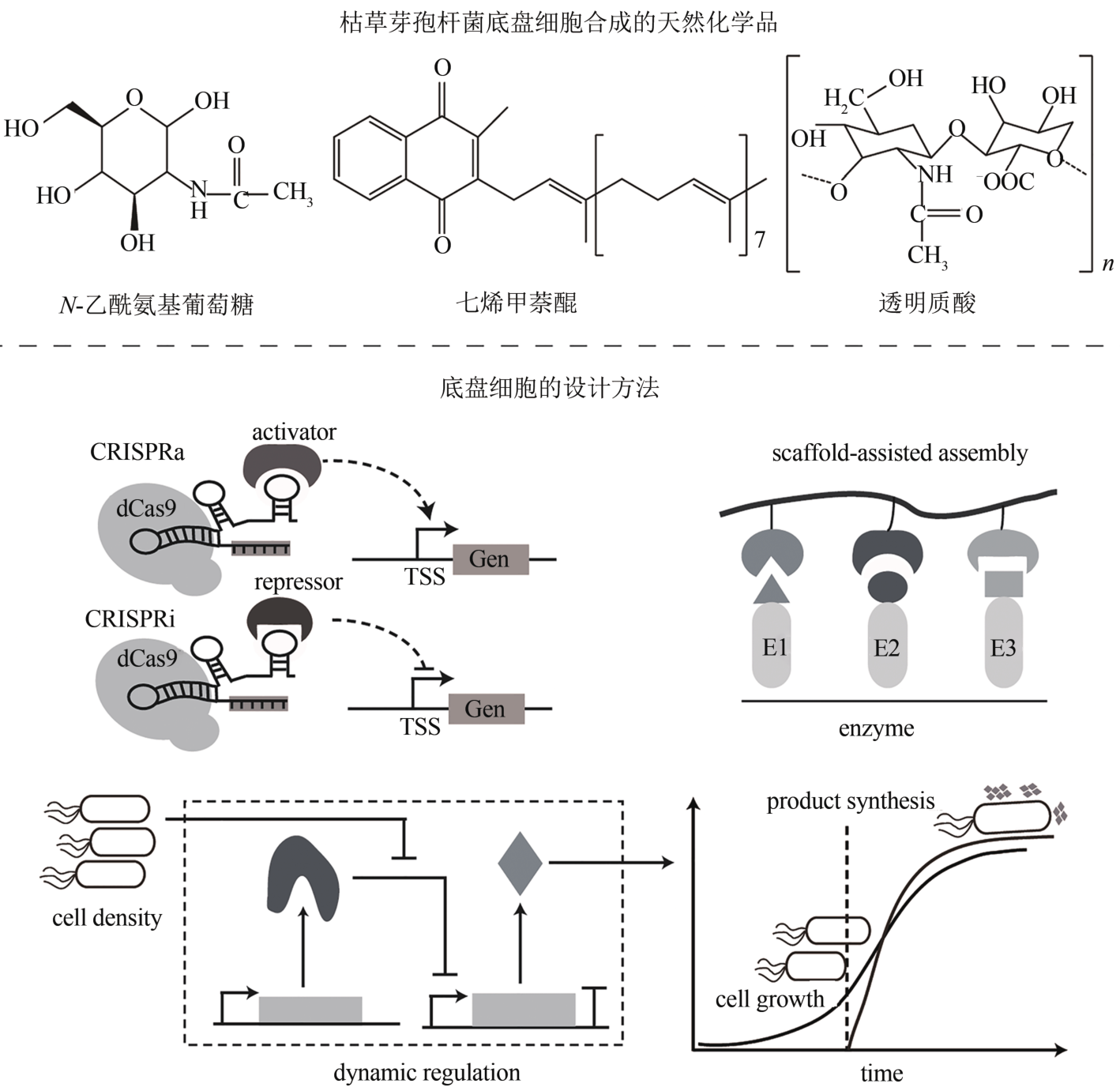
| 基因回路 | glmS核糖开关 | 基于GlcN6P响应的天然glmS核糖开关 | [ |
| 配体-适体生物传感器 | 基于配体凝血酶设计的双功能基因表达调控电路 | [ | |
| ADC系统 | 基于生物传感器、CRISPR和合成基因回路的代谢流量动态自发双重调控(autonomous dual control,ADC)系统 | [ | |
| 内源性调控系统 | Sec分泌途径 | 通过改善Sec分泌途径,提高异源蛋白的分泌 | [ |
| 群体感应系统 | 细胞密度达到阈值,动态调节相关基因的表达 | [ | |
| I型毒素-抗毒素系统 | 通过抗毒素反义RNA与毒素mRNA互补配对,下调基因的表达 | [ | |
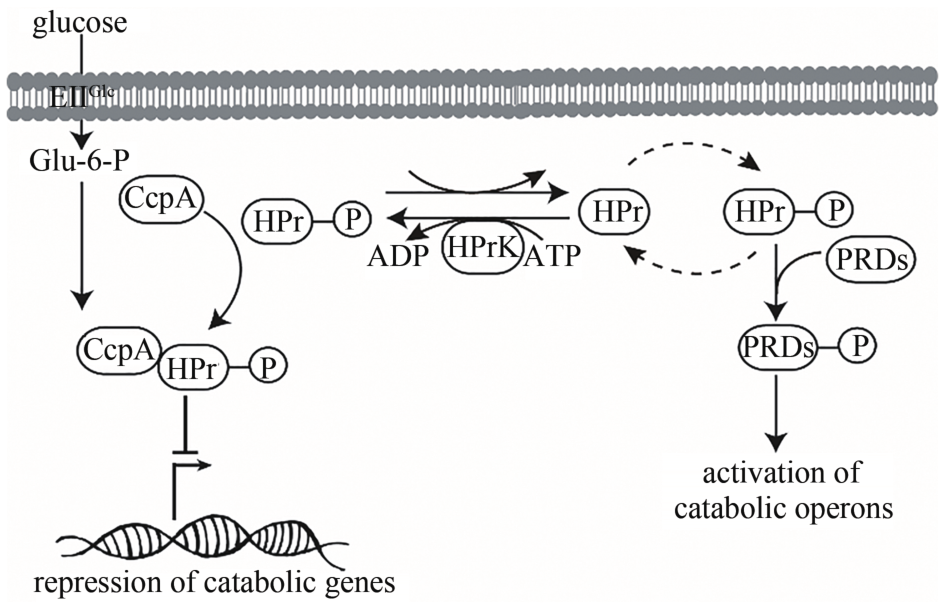
| 碳分解代谢阻遏作用 | 解除碳分解代谢阻遏作用,细胞可同时利用多种碳源 | [ |
Tab. 1 Methods for the construction of chassis cell in B. subtilis
| 类别 | 构建方法 | 功能特性 | 文献 |
|---|---|---|---|
| 基因编辑 | 基因组简化 | 利用无痕基因敲除技术构建多种基因组简化菌株 | [ |
| CRISPR/Cas9迭代系统 | 实现基因敲除、点突变和基因敲入,避免脱靶效应 | [ | |
| CRISPR-Cas9工具包 | 实现基因敲除、多基因敲入和转录抑制 | [ | |
| CAMERS-B系统 | 通过设计crRNA阵列实现多基因编辑和调控 | [ | |
| 内源性启动子库 | 基于细胞生长构建的内源性启动子文库 | [ | |
| 模块化操作单元 | 在转录翻译和蛋白水解水平上共同调节基因和蛋白的表达 | [ | |
| DNA支架 | 通过DNA支架调节途径酶的空间比例和方向 | [ | |
| RNA支架 | 通过RNA支架与特异性蛋白结合,转录抑制靶基因的表达 | [ | |
| 内源性空间支架 | 通过FMMs空间支架构建FMMs-多酶复合物系统 | [ | |
| 无细胞蛋白表达系统 | 以外源DNA为模板,利用细胞裂解物,可在体外表达蛋白质 | [ | |
| 基因回路 | glmS核糖开关 | 基于GlcN6P响应的天然glmS核糖开关 | [ |
| 配体-适体生物传感器 | 基于配体凝血酶设计的双功能基因表达调控电路 | [ | |
| ADC系统 | 基于生物传感器、CRISPR和合成基因回路的代谢流量动态自发双重调控(autonomous dual control,ADC)系统 | [ | |
| 内源性调控系统 | Sec分泌途径 | 通过改善Sec分泌途径,提高异源蛋白的分泌 | [ |
| 群体感应系统 | 细胞密度达到阈值,动态调节相关基因的表达 | [ | |
| I型毒素-抗毒素系统 | 通过抗毒素反义RNA与毒素mRNA互补配对,下调基因的表达 | [ | |
| 碳分解代谢阻遏作用 | 解除碳分解代谢阻遏作用,细胞可同时利用多种碳源 | [ |
| 名称 | 类型 | 性质 | 文献 |
|---|---|---|---|
| P43 | 组成型 | B. subtilis中cdd基因的启动子,能持续性高效表达基因 | [ |
| P HpaII | 组成型 | 来源于Staphylococcus aureus中的载体pUB110 | [ |
| P laps | 组成型 | 人工构建的双启动子,启动强度是P43的13倍 | [ |
| P grac | 诱导型 | IPTG诱导,由lac操纵子和groESL启动子融合而成 | [ |
| P xyl | 诱导型 | 受木糖诱导,且受葡萄糖抑制 | [ |
| P sacB | 诱导型 | 受蔗糖诱导 | [ |
| P mtlA | 诱导型 | 受甘露醇诱导 | [ |
| P glv | 诱导型 | 受麦芽糖诱导 | [ |
| P amy | 诱导型 | 受淀粉诱导,可以抵抗碳分解代谢物阻遏的启动子 | [ |
| P ohrB | 诱导型 | 受环境压力和葡萄糖匮乏诱导 | [ |
| P gsiB | 诱导性 | 受环境因素(热、盐、酸、乙醇、缺氧等)诱导 | [ |
| P rpsF | 时期特异型 | 在对数期被激活 | [ |
| P srfA | 时期特异型 | 在对数中后期被激活 | [ |
| P aprE | 时期特异型 | 在对数末期被激活 | [ |
| P cry3Aa | 时期特异型 | 在稳定期被激活 | [ |
| P manP | 自诱导型 | 适合于高密度发酵 | [ |
| P bs | 合成型 | 在E.coli、B. subtilis和S. cerevisiae均有活力的广谱启动子 | [ |
Tab. 2 Common promoters for the B. subtilis expression system
| 名称 | 类型 | 性质 | 文献 |
|---|---|---|---|
| P43 | 组成型 | B. subtilis中cdd基因的启动子,能持续性高效表达基因 | [ |
| P HpaII | 组成型 | 来源于Staphylococcus aureus中的载体pUB110 | [ |
| P laps | 组成型 | 人工构建的双启动子,启动强度是P43的13倍 | [ |
| P grac | 诱导型 | IPTG诱导,由lac操纵子和groESL启动子融合而成 | [ |
| P xyl | 诱导型 | 受木糖诱导,且受葡萄糖抑制 | [ |
| P sacB | 诱导型 | 受蔗糖诱导 | [ |
| P mtlA | 诱导型 | 受甘露醇诱导 | [ |
| P glv | 诱导型 | 受麦芽糖诱导 | [ |
| P amy | 诱导型 | 受淀粉诱导,可以抵抗碳分解代谢物阻遏的启动子 | [ |
| P ohrB | 诱导型 | 受环境压力和葡萄糖匮乏诱导 | [ |
| P gsiB | 诱导性 | 受环境因素(热、盐、酸、乙醇、缺氧等)诱导 | [ |
| P rpsF | 时期特异型 | 在对数期被激活 | [ |
| P srfA | 时期特异型 | 在对数中后期被激活 | [ |
| P aprE | 时期特异型 | 在对数末期被激活 | [ |
| P cry3Aa | 时期特异型 | 在稳定期被激活 | [ |
| P manP | 自诱导型 | 适合于高密度发酵 | [ |
| P bs | 合成型 | 在E.coli、B. subtilis和S. cerevisiae均有活力的广谱启动子 | [ |
| 类别 | 产品 | 菌株 | 方法 | 产量 | 文献 |
|---|---|---|---|---|---|
| 生物化学品 | 七烯甲萘醌 | B. subtilis | Phr60-Rap60-Spo0A群体响应调控系统 | 360 mg/L | [ |
| 核黄素 | B. subtilis RH33 | 定点诱变zwf、gnd | 15.7 g/L | [ | |
| 鲨肌醇 | B. subtilis KU303 | 敲除iolABCDEF、iolHIJ、iolX、iolR,过表达IolG、IolW、IolT、PntAB | 27.6 g/L | [ | |
| 软骨素 | B. subtilis E168H | 表达kfoC-kfoA、kfiC-kfiA,上调tuaD | 5.22 g/L | [ | |
| 鸟苷 | BSK814G2 | 敲除purA被敲除,共表达PRS、purF、guaB | 115.2 mg/L | [ | |
| 胸苷 | BSK756T3 | 敲除tdk,过表达prs、ushA、thyA、dut、ndk | 151.2 mg/L | [ | |
| 紫穗槐二烯 | B. subtilis 1A1 | 过表达dxs、idi、ads | 20 mg/L | [ | |
| 乙缩醛 | B. subtilis BMN | 敲除bdhA,过表达基因 nox | 56.7 g/L | [ | |
| 2,3-丁二醇 | B. subtilis BSF9 | 敲除upp、acoA、bdhA、pta、ldh,过表达alsS、alsD、budC、udhA | 103.7 g/L | [ | |
| 异丁醇 | BSUL08 | 敲除alsS、ldh、pdhC、pgi,过表达zwf、udhA | 6.12 g/L | [ | |
| 二吡啶甲酸 | B. subtilis OA105 | 敲除alsSD,过表达spoVGBA | 2.3 g/L | [ | |
| 苹果酸 | BSUPML | 敲除ldh,过表达异源基因mdh | 1.23 g/L | [ | |
| 透明质酸 | B. subtilis E168TH | 表达hasA、tuaD、gtaB、glmU、glmM、glmS | 19.38 g/L | [ | |
| N-乙酰氨基葡萄糖 | BP18-afGNA1 | 重构中心碳和氧化还原代谢 | 24.5 g/L | [ | |
| 典型化学品 | α-淀粉酶 | B. subtilis 1A237 | 共表达PrsA和DnaK蛋白 | 1352 U/mL | [ |
| 纳豆激酶 | BSG328 | 改造srfA启动子核心区 | 93.6 U/mL | [ | |
| 氨肽酶 | BSG329 | 改造srfA启动子核心区 | 69.8 U/mL | [ | |
| L-天冬酰胺酶 | B. subtilis WB600 | 表达type Ⅱ ASN | 407.6 U/mL | [ | |
| β-环糊精糖基转移酶 | BS5 ATCC 6051a | 敲除srfC、spoIIAC、nprE、aprE、amyE | 227.8 U/mL | [ | |
| 植酸酶 | B. subtilis BD170 | 表达phyC | 47.7 U/mL | [ |
Tab. 3 Typical bio-chemicals and industrial enzymes produced by B. subtilis chassis cell
| 类别 | 产品 | 菌株 | 方法 | 产量 | 文献 |
|---|---|---|---|---|---|
| 生物化学品 | 七烯甲萘醌 | B. subtilis | Phr60-Rap60-Spo0A群体响应调控系统 | 360 mg/L | [ |
| 核黄素 | B. subtilis RH33 | 定点诱变zwf、gnd | 15.7 g/L | [ | |
| 鲨肌醇 | B. subtilis KU303 | 敲除iolABCDEF、iolHIJ、iolX、iolR,过表达IolG、IolW、IolT、PntAB | 27.6 g/L | [ | |
| 软骨素 | B. subtilis E168H | 表达kfoC-kfoA、kfiC-kfiA,上调tuaD | 5.22 g/L | [ | |
| 鸟苷 | BSK814G2 | 敲除purA被敲除,共表达PRS、purF、guaB | 115.2 mg/L | [ | |
| 胸苷 | BSK756T3 | 敲除tdk,过表达prs、ushA、thyA、dut、ndk | 151.2 mg/L | [ | |
| 紫穗槐二烯 | B. subtilis 1A1 | 过表达dxs、idi、ads | 20 mg/L | [ | |
| 乙缩醛 | B. subtilis BMN | 敲除bdhA,过表达基因 nox | 56.7 g/L | [ | |
| 2,3-丁二醇 | B. subtilis BSF9 | 敲除upp、acoA、bdhA、pta、ldh,过表达alsS、alsD、budC、udhA | 103.7 g/L | [ | |
| 异丁醇 | BSUL08 | 敲除alsS、ldh、pdhC、pgi,过表达zwf、udhA | 6.12 g/L | [ | |
| 二吡啶甲酸 | B. subtilis OA105 | 敲除alsSD,过表达spoVGBA | 2.3 g/L | [ | |
| 苹果酸 | BSUPML | 敲除ldh,过表达异源基因mdh | 1.23 g/L | [ | |
| 透明质酸 | B. subtilis E168TH | 表达hasA、tuaD、gtaB、glmU、glmM、glmS | 19.38 g/L | [ | |
| N-乙酰氨基葡萄糖 | BP18-afGNA1 | 重构中心碳和氧化还原代谢 | 24.5 g/L | [ | |
| 典型化学品 | α-淀粉酶 | B. subtilis 1A237 | 共表达PrsA和DnaK蛋白 | 1352 U/mL | [ |
| 纳豆激酶 | BSG328 | 改造srfA启动子核心区 | 93.6 U/mL | [ | |
| 氨肽酶 | BSG329 | 改造srfA启动子核心区 | 69.8 U/mL | [ | |
| L-天冬酰胺酶 | B. subtilis WB600 | 表达type Ⅱ ASN | 407.6 U/mL | [ | |
| β-环糊精糖基转移酶 | BS5 ATCC 6051a | 敲除srfC、spoIIAC、nprE、aprE、amyE | 227.8 U/mL | [ | |
| 植酸酶 | B. subtilis BD170 | 表达phyC | 47.7 U/mL | [ |
| 1 | LEE J W, NA D, PARK J M,et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals [J]. Nature Chemical Biology,2012,8(6): 536-546. |
| 2 | LEE S, MATTANOVICH D, VILLAVERDE A. Systems metabolic engineering,industrial biotechnology and microbial cell factories [J]. Microbial Cell Factories,2012,11(1): 156. |
| 3 | GOEL A, WORTEL M T, MOLENAAR D,et al. Metabolic shifts: a fitness perspective for microbial cell factories [J]. Biotechnology Letters,2012,34(12): 2147-2160. |
| 4 | LIU L, LIU Y, SHIN H D,et al. Developing Bacillus spp. as a cell factory for production of microbial enzymes and industrially important biochemicals in the context of systems and synthetic biology [J]. Applied Microbiology and Biotechnology,2013,97(14): 6113-6127. |
| 5 | JUHAS M,REU D R, ZHU B,et al. Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering [J]. Microbiology,2014,160(11): 2341-2351. |
| 6 | 李杨. 基因组简化枯草芽孢杆菌的构建及应用 [D]. 天津:天津大学,2017. |
| LI Y. Construction of genome reduced Bacillus subtilis and their application in production of bio-based chemicals [D]. Tianjin: Tianjin University,2017. | |
| 7 | SO Y, PARK S Y, PARK E H,et al. A highly efficient CRISPR-Cas9-Mediated large genomic deletion in Bacillus subtilis [J]. Frontiers in Microbiology,2017,8: a1167. |
| 8 | WESTBROOK A W, YOUNG M, CHOU C P. Development of a CRISPR-Cas9 tool kit for comprehensive engineering of Bacillus subtilis [J]. Applied and Environmental Microbiology,2016,82(16): 4876-4895. |
| 9 | WU Y, LIU Y, LV X,et al. CAMERS-B: CRISPR/Cpf1 Assisted multiple-genes editing and regulation system for Bacillus subtilis [J]. Biotechnology and Bioengineering,2020. DOI: org/10.1002/bit.27322 . |
| 10 | SONG Y, NIKOLOFF J M, FU G,et al. Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications [J]. PLoS One,2016,11(7): E158447. |
| 11 | GUIZIOU S, SAUVEPLANE V, CHANG H J,et al. A part toolbox to tune genetic expression in Bacillus subtilis [J]. Nucleic Acids Research,2016,44(15): 7495-7508. |
| 12 | LIU Y, ZHU Y, MA W,et al. Spatial modulation of key pathway enzymes by DNA-guided scaffold system and respiration chain engineering for improved N-acetylglucosamine production by Bacillus subtilis [J]. Metabolic Engineering,2014,24: 61-69. |
| 13 | LIU Y, ZHU Y, LI J,et al. Modular pathway engineering of Bacillus subtilis for improved N-acetylglucosamine production [J]. Metabolic Engineering,2014,23: 42-52. |
| 14 | LV X, WU Y, TIAN R,et al. Synthetic metabolic channel by functional membrane microdomains for compartmentalized flux control [J]. Metabolic Engineering,2020,59: 106-118. |
| 15 | KELWICK R, WEBB A J, MACDONALD J T,et al. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements [J]. Metabolic Engineering,2016,38: 370-381. |
| 16 | NIU T, LIU Y, LI J,et al. Engineering a glucosamine-6-phosphate responsive glmS ribozyme switch enables dynamic control of metabolic flux in Bacillus subtilis for overproduction of N-acetylglucosamine [J]. ACS Synthetic Biology,2018,7(10): 2423-2435. |
| 17 | DENG J, CHEN C, GU Y,et al. Creating an in vivo bifunctional gene expression circuit through an aptamer-based regulatory mechanism for dynamic metabolic engineering in Bacillus subtilis [J]. Metabolic Engineering,2019,55: 179-190. |
| 18 | WU Y, CHEN T, LIU Y,et al. Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis [J]. Nucleic Acids Research,2020,48(2): 996-1009. |
| 19 | CHEN J, FU G, GAI Y,et al. Combinatorial Sec pathway analysis for improved heterologous protein secretion in Bacillus subtilis: identification of bottlenecks by systematic gene overexpression [J]. Microbial Cell Factories,2015,14(1): 92. |
| 20 | FERREIRA L C S, FERREIRA R C C, SCHUMANN W. Bacillus subtilis as a tool for vaccine development: from antigen factories to delivery vectors [J]. Anais Da Academia Brasileira De Ciências,77(1): 113-124. |
| 21 | GUAN C, CUI W, CHENG J,et al. Development of an efficient autoinducible expression system by promoter engineering in Bacillus subtilis [J]. Microbial Cell Factories,2016,15(1): 66. |
| 22 | CUI S, LV X, WU Y,et al. Engineering a bifunctional Phr60-Rap60-Spo0A quorum-sensing molecular switch for dynamic fine-tuning of menaquinone-7 synthesis in Bacillus subtilis [J]. ACS Synthetic Biology,2019,8(8): 1826-1837. |
| 23 | YANG S, WANG Y, WEI C,et al. A new sRNA-mediated posttranscriptional regulation system for Bacillus subtilis [J]. Biotechnology and Bioengineering,2018,115(12): 2986-2995. |
| 24 | REU D R, RATH H, RMER A TH,et al. Changes of DNA topology affect the global transcription landscape and allow rapid growth of a Bacillus subtilis mutant lacking carbon catabolite repression [J]. Metabolic Engineering,2018,45: 171-179. |
| 25 | ARA K, OZAKI K, NAKAMURA K,et al. Bacillus minimum genome factory: effective utilization of microbial genome information [J]. Biotechnology and Applied Biochemistry,2007,46(3): 169-178. |
| 26 | WESTERS H, DORENBOS R, DIJL J M VAN,et al. Genome engineering reveals large dispensable regions in Bacillus subtilis [J]. Molecular Biology and Evolution,2003,20(12): 2076-2090. |
| 27 | TAKUYA M, RYOSUKE K, KEIJI E,et al. Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis [J]. DNA Research,2008,15(2): 73-81. |
| 28 | JAKOČIŪNAS T, JENSEN M K, KEASLING J D. CRISPR/Cas9 advances engineering of microbial cell factories [J]. Metabolic Engineering,2016,34: 44-59. |
| 29 | DOUDNA J A, CHARPENTIER E. The new frontier of genome engineering with CRISPR-Cas9 [J]. Science,2014,346(6213): 1258096. |
| 30 | BIKARD D, JIANG W, SAMAI P,et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system [J]. Nucleic Acids Research,2013,41(15): 7429-7437. |
| 31 | JEONG D E, PARK S H, PAN J G,et al. Genome engineering using a synthetic gene circuit in Bacillus subtilis [J]. Nucleic Acids Research,2014,43(6): e42. |
| 32 | ISAACS F J, DWYER D J, DING C,et al. Engineered riboregulators enable post-transcriptional control of gene expression [J]. Nature Biotechnology,2004,22(7): 841-847. |
| 33 | KIM J Y H,CHA H J. Down-regulation of acetate pathway through antisense strategy in Escherichia coli: improved foreign protein production [J]. Biotechnology and Bioengineering,2003,83(7): 841-853. |
| 34 | MAN S, CHENG R, MIAO C,et al. Artificial trans-encoded small non-coding RNAs specifically silence the selected gene expression in bacteria [J]. Nucleic Acids Research,2011,39(8): e50. |
| 35 | DOMINGUEZ A A,LIM W A, QI L S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation [J]. Nature Reviews Molecular Cell Biology,2016,17(1): 5-15. |
| 36 | DONG C, FONTANA J, PATEL A,et al. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria [J]. Nature Communications,2018,9(1): 2489. |
| 37 | ZHU H, LIANG C. CRISPR D T: designing gRNAs for the CRISPR-Cpf1 system with improved target efficiency and specificity [J]. Bioinformatics,2019,35(16): 2783-2789. |
| 38 | ZHANG X Z, CUI Z L, HONG Q,et al. High level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800 [J]. Applied and Environmental Microbiology,2005,71(7): 4101. |
| 39 | ZYPRIAN E V A, MATZURA H. Characterization of signals promoting gene expression on the Staphylococcus aureus plasmid pUB110 and development of a gram-positive expression vector system [J]. DNA,1986,5(3): 219-225. |
| 40 | YANG M, ZHANG W, JI S,et al. Generation of an artificial double promoter for protein expression in Bacillus subtilis through a promoter trap system [J]. PLoS One,2013,8(2): E56321. |
| 41 | PHAN T T P, NGUYEN H D, SCHUMANN W. Development of a strong intracellular expression system for Bacillus subtilis by optimizing promoter elements [J]. Journal of Biotechnology,2012,157(1): 167-172. |
| 42 | KIM L, MOGK A, SCHUMANN W. A xylose-inducible Bacillus subtilis integration vector and its application [J]. Gene,1996,181(1): 71-76. |
| 43 | YE R, KIM J H, KIM B G,et al. High-level secretory production of intact,biologically active staphylokinase from Bacillus subtilis [J]. Biotechnology and Bioengineering,1999,62(1): 87-96. |
| 44 | MORABBI H K, WENZEL M, ALTENBUCHNER J. Regulation of mtl operon promoter of Bacillus subtilis: requirements of its use in expression vectors [J]. Microbial Cell Factories,2011,10(1): 83. |
| 45 | MING Y M, WEI Z W, LIN C Y,et al. Development of a Bacillus subtilis expression system using the improved P glv promoter [J]. Microbial Cell Factories,2010,9(1): 55. |
| 46 | NAGARAJAN D R, KRISHNAN C. Use of a new catabolite repression resistant promoter isolated from Bacillus subtilis KCC103 for hyper-production of recombinant enzymes [J]. Protein Expression and Purification,2010,70(1): 122-128. |
| 47 | PANAHI R, VASHEGHANI-FARAHANI E, SHOJAOSADATI S A,et al. Induction of Bacillus subtilis expression system using environmental stresses and glucose starvation [J]. Annals of Microbiology,2014,64(2): 879-882. |
| 48 | MITTENHUBER G. A phylogenomic study of the general stress response sigma factor sigmaB of Bacillus subtilis and its regulatory proteins [J]. J. Mol. Microbiol. Biotechnol.,2002,4(4): 427-452. |
| 49 | NIJLAND R, LINDNER C, HARTSKAMP M VAN,et al. Heterologous production and secretion of Clostridium perfringens β-toxoid in closely related Gram-positive hosts [J]. Journal of Biotechnology,2007,127(3): 361-372. |
| 50 | WILLENBACHER J, MOHR T, HENKEL M,et al. Substitution of the native srfA promoter by constitutive P veg in two B. subtilis strains and evaluation of the effect on Surfactin production [J]. Journal of Biotechnology,2016,224: 14-17. |
| 51 | OGURA M, SHIMANE K, ASAI K,et al. Binding of response regulator DegU to the aprE promoter is inhibited by RapG,which is counteracted by extracellular PhrG in Bacillus subtilis [J]. Molecular Microbiology,2003,49: 1685-1697. |
| 52 | LEE S J, PAN J G, PARK S H,et al. Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis [J]. Journal of Biotechnology,2010,149(1): 16-20. |
| 53 | WENZEL M, LLER A, SIEMANN H M,et al. Self-inducible Bacillus subtilis expression system for reliable and inexpensive protein production by high-cell-density fermentation [J]. Applied and Environmental Microbiology,2011,77(18): 6419. |
| 54 | YANG S, LIU Q, ZHANG Y,et al. Construction and characterization of broad-spectrum promoters for synthetic biology [J]. ACS Synthetic Biology,2018,7(1): 287-291. |
| 55 | SIU K H, CHEN R P, SUN Q,et al. Synthetic scaffolds for pathway enhancement [J]. Current Opinion in Biotechnology,2015,36: 98-106. |
| 56 | CHANDRASEKARAN A R. Programmable DNA scaffolds for spatially-ordered protein assembly [J]. Nanoscale,2016,8(8): 4436-4446. |
| 57 | PORTER E B, POLASKI J T, MORCK M M,et al. Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors [J]. Nature Chemical Biology,2017,13(3): 295-301. |
| 58 | CONRADO R J, WU G C, BOOCK J T,et al. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency [J]. Nucleic Acids Research,2011,40(4): 1879-1889. |
| 59 | DELEBECQUE C J, SILVER P A, LINDNER A B. Designing and using RNA scaffolds to assemble proteins in vivo [J]. Nature Protocols,2012,7(10): 1797-1807. |
| 60 | KONERMANN S, BRIGHAM M D, TREVINO A E,et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex [J]. Nature,2015,517(7536): 583-588. |
| 61 | PFEIFFER V, PAPENFORT K, LUCCHINI S,et al. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation [J]. Nature Structural and Molecular Biology,2009,16(8): 840-846. |
| 62 | LEE M J, MANTELL J, HODGSON L,et al. Engineered synthetic scaffolds for organizing proteins within the bacterial cytoplasm [J]. Nature Chemical Biology,2018,14(2): 142-147. |
| 63 | BRAMKAMP M, LOPEZ D. Exploring the existence of lipid rafts in bacteria [J]. Microbiology and Molecular Biology Reviews,2015,79(1): 81. |
| 64 | LV X, JIN K, WU Y,et al. Enzyme assembly guided by SPFH-induced functional inclusion bodies for enhanced cascade biocatalysis [J]. Biotechnology and Bioengineering, 2020. DOI: org/10. 1002/bit.27304 . |
| 65 | ANESIADIS N, KOBAYASHI H, CLUETT W R,et al. Analysis and design of a genetic circuit for dynamic metabolic engineering [J]. ACS Synthetic Biology,2013,2(8): 442-452. |
| 66 | ZHANG J, JENSEN M K, KEASLING J D. Development of biosensors and their application in metabolic engineering [J]. Current Opinion in Chemical Biology,2015,28:1-8. |
| 67 | MANDAL M, BREAKER R R. Gene regulation by riboswitches [J]. Nature Reviews Molecular Cell Biology,2004,5(6): 451-463. |
| 68 | BERENS C, GROHER F, SUESS B. RNA aptamers as genetic control devices: the potential of riboswitches as synthetic elements for regulating gene expression [J]. Biotechnology Journal,2015,10(2): 246-257. |
| 69 | PEREZ J G, STARK J C, JEWETT M C. Cell-free synthetic biology: engineering beyond the cell [J]. Cold Spring Harbor Perspectives in Biology,2016,8(12): A023853. |
| 70 | HONG S H, KWON Y C, JEWETT M C. Non-standard amino acid incorporation into proteins using Escherichia coli cell-free protein synthesis [J]. Frontiers in Chemistry,2014,2: A34. |
| 71 | SHRESTHA P, HOLLAND T M, BUNDY B C. Streamlined extract preparation for Escherichia coli based cell-free protein synthesis by sonication or bead vortex mixing [J]. BioTechniques,2012,53(3): 163-174. |
| 72 | KARIM A S, JEWETT M C. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery [J]. Metabolic Engineering,2016,36: 116-126. |
| 73 | NGUYEN H D, PHAN T T P, SCHUMANN W. Analysis and application of Bacillus subtilis sortases to anchor recombinant proteins on the cell wall [J]. AMB Express,2011,1(1): 22. |
| 74 | CASCHERA F, NOIREAUX V. A cost-effective polyphosphate-based metabolism fuels an all Escherichia coli cell-free expression system [J]. Metabolic Engineering,2015,27: 29-37. |
| 75 | SONG Y, NIKOLOFF J M, ZHANG D. Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus subtilis [J]. Journal of Microbiology Biotechnology,2015,25(7): 963-977. |
| 76 | WATERS C M, BASSLER B L. Quorum sensing: Cell-to-cell communication in bacteria [J]. Annual Review of Cell and Developmental Biology,2005,21(1): 319-346. |
| 77 | MILLER M B, BASSLER B L. Quorum sensing in bacteria [J]. Annual Review of Microbiology,2001,55(1): 165-199. |
| 78 | LÓPEZ D, KOLTER R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis [J]. FEMS Microbiol Review,2010,34(2): 134-149. |
| 79 | SCHULTZ D, WOLYNES P G, JACOB E B,et al. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis [J]. PNAS,2009,106(50): 21027-21034. |
| 80 | HAMOEN L W, VENEMA G, KUIPERS O P. Controlling competence in Bacillus subtilis: shared use of regulators [J]. Microbiology,2003,149(1): 9-17. |
| 81 | SHANK E A, KOLTER R. Extracellular signaling and multicellularity in Bacillus subtilis [J]. Current Opinion in Microbiology,2011,14(6): 741-747. |
| 82 | BOGUSLAWSKI K M, HILL P A, GRIFFITH K L. Novel mechanisms of controlling the activities of the transcription factors Spo0A and ComA by the plasmid-encoded quorum sensing regulators Rap60-Phr60 in Bacillus subtilis [J]. Molecular Microbiology,2015,96(2): 325-348. |
| 83 | DURAND S, JAHN N, CONDON C,et al. Type I toxin-antitoxin systems in Bacillus subtilis [J]. RNA Biology,2012,9(12): 1491-1497. |
| 84 | JAHN N, PREIS H, WIEDEMANN C,et al. BsrG/SR4 from Bacillus subtilis-the first temperature dependent type I toxin-antitoxin system [J]. Molecular Microbiology,2012,83(3): 579-598. |
| 85 | GÖRKE B, STÜLKE J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients [J]. Nature Reviews Microbiology,2008,6(8): 613-624. |
| 86 | SINGH K D, SCHMALISCH M H, STÜLKE J,et al. Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources [J]. Journal of Bacteriology,2008,190(21): 7275-7284. |
| 87 | 应明,班睿. 枯草芽孢杆菌ccpA基因敲除及对其核黄素产量的影响[J].微生物学报,2006,46(1):23-27 |
| YING M, BAN R. Knockout of the ccpA gene in Bacillus subtilis and influence on riboflavin production [J]. Acta Microbiologica Sinica,2006,46(1): 23-27. | |
| 88 | WANG Z, CHEN T, MA X,et al. Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf and gnd gene from Corynebacterium glutamicum [J]. Bioresource Technology,2011,102(4): 3934-3940. |
| 89 | TANAKA K, TAKANAKA S, YOSHIDA K I. A second-generation Bacillus cell factory for rare inositol production [J]. Bioengineered,2014,5(5): 331-334. |
| 90 | JIN P, ZHANG L, YUAN P,et al. Efficient biosynthesis of polysaccharides chondroitin and heparosan by metabolically engineered Bacillus subtilis [J]. Carbohydrate Polymers,2016,140: 424-432. |
| 91 | FENG Y, LIU S, JIAO Y,et al. Enhanced extracellular production of L-asparaginase from Bacillus subtilis 168 by B. subtilis WB600 through a combined strategy [J]. Applied Microbiology and Biotechnology,2017,101(4): 1509-1520. |
| 92 | LI Y, ZHU X, ZHANG X,et al. Characterization of genome-reduced Bacillus subtilis strains and their application for the production of guanosine and thymidine [J]. Microbial Cell Factories,2016,15(1): 94. |
| 93 | ZHOU K, ZOU R, ZHANG C,et al. Optimization of amorphadiene synthesis in Bacillus subtilis via transcriptional,translational,and media modulation [J]. Biotechnology and Bioengineering,2013,110(9): 2556-2561. |
| 94 | ZHANG X, ZHANG R, BAO T,et al. The rebalanced pathway significantly enhances acetoin production by disruption of acetoin reductase gene and moderate-expression of a new water-forming NADH oxidase in Bacillus subtilis [J]. Metabolic Engineering,2014,23: 34-41. |
| 95 | FU J, HUO G, FENG L,et al. Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production [J]. Biotechnology for Biofuels,2016,9(1): 90. |
| 96 | QI H, LI S, ZHAO S,et al. Model-driven redox pathway manipulation for improved isobutanol production in Bacillus subtilis complemented with experimental validation and metabolic profiling analysis [J]. PLoS One,2014,9(4): e93815. |
| 97 | TOYA Y, HIRASAWA T, ISHIKAWA S,et al. Enhanced dipicolinic acid production during the stationary phase in Bacillus subtilis by blocking acetoin synthesis [J]. Bioscience,Biotechnology,and Biochemistry,2015,79(12): 2073-2080. |
| 98 | MU L, WEN J. Engineered Bacillus subtilis 168 produces L-malate by heterologous biosynthesis pathway construction and lactate dehydrogenase deletion [J]. World Journal of Microbiology and Biotechnology,2013,29(1): 33-41. |
| 99 | JIN P, KANG Z, YUAN P,et al. Production of specific-molecular-weight hyaluronan by metabolically engineered Bacillus subtilis 168 [J]. Metabolic Engineering,2016,35: 21-30. |
| 100 | ZHANG K, DUAN X, WU J. Multigene disruption in undomesticated Bacillus subtilis ATCC 6051a using the CRISPR/Cas9 system [J]. Scientific Reports,2016,6(1): 27943. |
| 101 | VUOLANTO A, WEYMARN N VON, KEROVUO J,et al. Phytase production by high cell density culture of recombinant Bacillus subtilis [J]. Biotechnology Letters,2001,23(10): 761-766. |
| 102 | GU Y, LV X, LIU Y,et al. Synthetic redesign of central carbon and redox metabolism for high yield production of N-acetylglucosamine in Bacillus subtilis [J]. Metabolic Engineering,2019,51: 59-69. |
| 103 | LIU Y, LIU L, SHIN H D,et al. Pathway engineering of Bacillus subtilis for microbial production of N-acetylglucosamine [J]. Metabolic Engineering,2013,19: 107-115. |
| 104 | LIU Y, LINK H, LIU L,et al. A dynamic pathway analysis approach reveals a limiting futile cycle in N-acetylglucosamine overproducing Bacillus subtilis [J]. Nature Communications,2016,7(1): 11933. |
| 105 | CUI S, XIA H, CHEN T,et al. Cell membrane and electron transfer engineering for improved synthesis of menaquinone-7 in Bacillus subtilis [J]. iScience,2020,23(3): 100918. |
| 106 | LIU S, HU W, WANG Z,et al. Production of riboflavin and related cofactors by biotechnological processes [J]. Microbial Cell Factories,2020,19(1): 31. |
| 107 | SHI S, SHEN Z, CHEN X,et al. Increased production of riboflavin by metabolic engineering of the purine pathway in Bacillus subtilis [J]. Biochemical Engineering Journal,2009,46(1): 28-33. |
| 108 | SHI T, WANG Y, WANG Z,et al. Deregulation of purine pathway in Bacillus subtilis and its use in riboflavin biosynthesis [J]. Microbial Cell Factories,2014,13(1): 101. |
| 109 | SHI S, CHEN T, ZHANG Z,et al. Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production [J]. Metabolic Engineering,2009,11(4): 243-252. |
| 110 | LIU L, LIU Y, LI J,et al. Microbial production of hyaluronic acid: current state,challenges,and perspectives [J]. Microbial Cell Factories,2011,10(1): 99. |
| 111 | WESTERS L, WESTERS H, QUAX W J. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism [J]. Biochimica Biophysica Acta,1694(3): 299-310. |
| [1] | CHEN Yingying, LIU Yang, SHI Junjie, MA Junying, JU Jianhua. CRISPR/Cas systems and their applications in gene editing with filamentous fungi [J]. Synthetic Biology Journal, 2024, 5(3): 672-693. |
| [2] | XU Zhimeng, XIE Zhen. Research progress and biotechnological applications of the prime editing [J]. Synthetic Biology Journal, 2024, 5(1): 1-15. |
| [3] | CHEN Yaru, CAO Yingxiu, SONG Hao. Advances and applications of gene editing and transcriptional regulation in electroactive microorganisms [J]. Synthetic Biology Journal, 2023, 4(6): 1281-1299. |
| [4] | SUN Meili, WANG Kaifeng, LU Ran, JI Xiaojun. Rewiring and application of Yarrowia lipolytica chassis cell [J]. Synthetic Biology Journal, 2023, 4(4): 779-807. |
| [5] | LIN Jicong, ZOU Gen, LIU Hongmin, WEI Yongjun. Application of CRISPR/Cas genome editing technology in the synthesis of secondary metabolites of filamentous fungi [J]. Synthetic Biology Journal, 2023, 4(4): 738-755. |
| [6] | LIU Ke, LIN Guihong, LIU Kun, ZHOU Wei, WANG Fengqing, WEI Dongzhi. Mining, engineering and functional expansion of CRISPR/Cas systems [J]. Synthetic Biology Journal, 2023, 4(1): 47-66. |
| [7] | LIANG Liya, LIU Rongming. Protein engineering of DNA targeting type Ⅱ CRISPR/Cas systems [J]. Synthetic Biology Journal, 2023, 4(1): 86-101. |
| [8] | LIU Jiaxin, CHENG Chi, LI Xinqi, WANG Chaojun, ZHANG Ying, XUE Chuang. Recent progress in the molecular genetic modification tools of Clostridium [J]. Synthetic Biology Journal, 2022, 3(6): 1201-1217. |
| [9] | LIU Qi, QIAN Zhilan, SONG Lili, YAO Chaoying, XU Mingqiang, REN Yanna, CAI Menghao. Rewiring and application of Pichia pastoris chassis cell [J]. Synthetic Biology Journal, 2022, 3(6): 1150-1173. |
| [10] | LIU Jiayu, YANG Zhihan, YANG Lei, ZHU Liying, ZHU Zhengming, JIANG Ling. Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnology [J]. Synthetic Biology Journal, 2022, 3(6): 1174-1200. |
| [11] | TAO Fei, SUN Tao, WANG Yu, WEI Ting, NI Jun, XU Ping. Challenges and opportunities in the research of Synechococcus chassis under the context of carbon peak and neutrality [J]. Synthetic Biology Journal, 2022, 3(5): 932-952. |
| [12] | ZHU Runtao, ZHONG Chao, DAI Zhuojun. Biofilm matrixes-from soft matters to engineered materials [J]. Synthetic Biology Journal, 2022, 3(4): 626-637. |
| [13] | SONG Fei, LIU Yuchen, CAI Zhiming, HUANG Weiren. Construction of tumor gene circuits using CRISPR/Cas tool and their applications [J]. Synthetic Biology Journal, 2022, 3(1): 53-65. |
| [14] | BI Jiacheng, TIAN Zhigang. Synthetic immunology and future NK cell immunotherapy [J]. Synthetic Biology Journal, 2022, 3(1): 22-34. |
| [15] | XIAO Han, LIU Yixin. Progress and challenge of the CRISPR-Cas system in gene editing for filamentous fungi [J]. Synthetic Biology Journal, 2021, 2(2): 274-286. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||