Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (1): 213-227.DOI: 10.12211/2096-8280.2024-046
• Research Article • Previous Articles
Effect of terminators on the downstream transcript unit with gene expression in Escherichiacoli
REN Jiawei1,2, ZHANG Jinpeng1,2, XU Guoqiang1,2, ZHANG Xiaomei3, XU Zhenghong4, ZHANG Xiaojuan1,2
- 1.Key Laboratory of Industrial Biotechnology of Ministry of Education,School of Biotechnology,Jiangnan University,Wuxi 214122,Jiangsu,China
2.National Engineering Research Center for Cereal Fermentation and Food Biomanufacturing,Jiangnan University,Wuxi 214122,Jiangsu,China
3.School of Life Science and Health Engineering,Jiangnan University,Wuxi 214122,Jiangsu,China
4.College of Light Industry Science and Engineering,Sichuan University,Chengdu 610065,Sichuan,China
-
Received:2024-06-17Revised:2024-08-22Online:2025-03-12Published:2025-02-28 -
Contact:ZHANG Xiaojuan
大肠杆菌中终止子对下游转录单元基因表达的影响
任家卫1,2, 张金鹏1,2, 徐国强1,2, 张晓梅3, 许正宏4, 张晓娟1,2
- 1.江南大学生物工程学院工业生物技术教育部重点实验室,江苏 无锡 214122
2.江南大学粮食发酵与食品生物制造国家工程研究中心,江苏 无锡 214122
3.江南大学生命科学与健康工程学院,江苏 无锡 214122
4.四川大学轻工科学与工程学院,四川 成都 610065
-
通讯作者:张晓娟 -
作者简介:任家卫 (1999—),男,硕士研究生。研究方向为终止子调控元件开发。E-mail:297887816@qq.com张晓娟 (1982—),女,教授。研究方向为酿造食品微生物开发与应用、新型工业菌株的基因表达调控元件开发。E-mail:zhangxj@jiangnan.edu.cn -
基金资助:国家自然科学基金(32171421)
CLC Number:
Cite this article
REN Jiawei, ZHANG Jinpeng, XU Guoqiang, ZHANG Xiaomei, XU Zhenghong, ZHANG Xiaojuan. Effect of terminators on the downstream transcript unit with gene expression in Escherichiacoli[J]. Synthetic Biology Journal, 2025, 6(1): 213-227.
任家卫, 张金鹏, 徐国强, 张晓梅, 许正宏, 张晓娟. 大肠杆菌中终止子对下游转录单元基因表达的影响[J]. 合成生物学, 2025, 6(1): 213-227.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-046
| Name | Description | Source |
|---|---|---|
| Escherichiacoli JM109 | EUROSCARF | |
| Escherichiacoli BL21 | EUROSCARF | |
| PTK | Dual Fluorescent Probe Plasmid,Kanr, T7 promoter, EGFP, mRFP1, T7 terminator | this study |
| PTK-pot | PTK derivatives carrying one-pot assembly libraries | this study |
| PTK-pheA-13-e | PTK derivatives carrying the terminator pheA, the spacer sequence sp-13, and the promoter dap-e | this study |
| PTK-pheA-13-e12 | PTK derivatives carrying the terminator pheA, the spacer sequence sp-13, and the promoter dap-e12 | this study |
| PTK-pheA-40-e | PTK derivatives carrying the terminator pheA, the spacer sequence sp-40, and the promoter dap-e | this study |
| PTK-pheA-40-e12 | PTK derivatives carrying the terminator pheA, the spacer sequence sp-40, and the promoter dap-e12 | this study |
| PTK-recA-13-e | PTK derivatives carrying the terminator recA, the spacer sequence sp-13 and the promoter dap-e | this study |
| PTK-recA-13-e12 | PTK derivatives carrying the terminator recA, the spacer sequence sp-13 and the promoter dap-e12 | this study |
| PTK-recA-40-e | PTK derivatives carrying the terminator recA, the spacer sequence sp-40 and the promoter dap-e | this study |
| PTK-recA-40-e12 | PTK derivatives carrying the terminator recA, the spacer sequence sp-40 and the promoter dap-e12 | this study |
Table 1 Strains and plasmids used in this study
| Name | Description | Source |
|---|---|---|
| Escherichiacoli JM109 | EUROSCARF | |
| Escherichiacoli BL21 | EUROSCARF | |
| PTK | Dual Fluorescent Probe Plasmid,Kanr, T7 promoter, EGFP, mRFP1, T7 terminator | this study |
| PTK-pot | PTK derivatives carrying one-pot assembly libraries | this study |
| PTK-pheA-13-e | PTK derivatives carrying the terminator pheA, the spacer sequence sp-13, and the promoter dap-e | this study |
| PTK-pheA-13-e12 | PTK derivatives carrying the terminator pheA, the spacer sequence sp-13, and the promoter dap-e12 | this study |
| PTK-pheA-40-e | PTK derivatives carrying the terminator pheA, the spacer sequence sp-40, and the promoter dap-e | this study |
| PTK-pheA-40-e12 | PTK derivatives carrying the terminator pheA, the spacer sequence sp-40, and the promoter dap-e12 | this study |
| PTK-recA-13-e | PTK derivatives carrying the terminator recA, the spacer sequence sp-13 and the promoter dap-e | this study |
| PTK-recA-13-e12 | PTK derivatives carrying the terminator recA, the spacer sequence sp-13 and the promoter dap-e12 | this study |
| PTK-recA-40-e | PTK derivatives carrying the terminator recA, the spacer sequence sp-40 and the promoter dap-e | this study |
| PTK-recA-40-e12 | PTK derivatives carrying the terminator recA, the spacer sequence sp-40 and the promoter dap-e12 | this study |
| Primer | Description |
|---|---|
| pheA-F | TAGCAACAATAAGGCCTCCCAAATCGGGGGGCCTTTTTTATTGAT |
| pheA-R | TTAGATCAATAAAAAAGGCCCCCCGATTTGGGAGGCCTTATTGTT |
| thrL-F | TAGCTCAAAAAAGCCCGCACCTGACAGTGCGGGCTTTTTTTTTACT |
| thrL-R | TTAGAGTAAAAAAAAAGCCCGCACTGTCAGGTGCGGGCTTTTTTGA |
| rpsO-F | TAGCCAGAAAAGGGGGCCTGAGTGGCCCCTTTTTTCAAGCT |
| rpsO-R | TTAGAGCTTGAAAAAAGGGGCCACTCAGGCCCCCTTTTCTG |
| arcA-F | TAGCAATAAAAACGGCGCTAAAAAGCGCCGTTTTTTTTGACG |
| arcA-R | TTAGCGTCAAAAAAAACGGCGCTTTTTAGCGCCGTTTTTATT |
| fhuE-F | TAGCGTAAAAAAGGCAGCCATCTGGCTGCCTTAGTCTCCCCA |
| fhuE-R | TTAGTGGGGAGACTAAGGCAGCCAGATGGCTGCCTTTTTTAC |
| hisI-F | TAGCCCCTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGT |
| hisI-R | TTAGACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGG |
| recA-F | TAGCAAGCAAAAGGGCCGCAGATGCGACCCTTGTGTATCAAC |
| recA-R | TTAGGTTGATACACAAGGGTCGCATCTGCGGCCCTTTTGCTT |
| T7-MOD-F | TAGCAAACAGATAGGCCCTCttcgGAGGGCCtatctgttTTTTTTT |
| T7-MOD-R | TTAGAAAAAAAaacagataGGCCCTCcgaaGAGGGCCTATCTGTTT |
| SP-TE-F | TAGCCCAATGTTTACTCATATCCAGTCACAGAAACTGAACTATC |
| SP-TE-R | TTAGGATAGTTCAGTTTCTGTGACTGGATATGAGTAAACATTGG |
| SPACE-5-F | CTAACTATC |
| SPACE-5-R | TTATGATAG |
| SPACE-13-F | CTAAAAACTGAACTATC |
| SPACE-13-R | TTATGATAGTTCAGTTT |
| SPACE-20-F | CTAAGTCACAGAAACTGAACTATC |
| SPACE-20-R | TTATGATAGTTCAGTTTCTGTGAC |
| SPACE-28-F | CTAACATATCCAGTCACAGAAACTGAACTATC |
| SPACE-28-R | TTATGATAGTTCAGTTTCTGTGACTGGATATG |
| SPACE-35-F | CTAAGTTTACTCATATCCAGTCACAGAAACTGAACTATC |
| SPACE-35-R | TTATGATAGTTCAGTTTCTGTGACTGGATATGAGTAAAC |
| SPACE-40-F | CTAACCAATGTTTACTCATATCCAGTCACAGAAACTGAACTATC |
| SPACE-40-R | TTATGATAGTTCAGTTTCTGTGACTGGATATGAGTAAACATTGG |
| dap-A16-F | ATAATTGTTTAACCCCCAAATGAGGGAAGAAGGTATAATTGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dap-A16-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCAATTATACCTTCTTCCCTCATTTGGGGGTTAAACAA |
| dap-e-F | ATAATTGTTTAGCCACCAAATGAGGGAAAGAGGCACAATGGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dap-e-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCCATTGTGCCTCTTTCCCTCATTTGGTGGCTAAACAA |
| dapA-e10-F | ATAATTGTTTTGACACCAAATGAGGGAATGTGGTATAATTGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dapA-e10-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCAATTATACCACATTCCCTCATTTGGTGTCAAAACAA |
| dapA-e11-F | ATAATTGTTTTGACACCAAATGAGGGAATGTGCTATAATGGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dapA-e11-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCCATTATAGCACATTCCCTCATTTGGTGTCAAAACAA |
| dapA-e12-F | ATAATTGTTTTGACACCAAATGAGGGAATGTGGTAGAGTGGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dapA-e12-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCCACTCTACCACATTCCCTCATTTGGTGTCAAAACAA |
| dapA-e10-35-F | ATAATTGTTTAACCCCCAAATGAGGGAATGTGGTATAATTGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dapA-e10-35-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCAATTATACCACATTCCCTCATTTGGGGGTTAAACAA |
| J1-F | ATAATTGACAATTTTCTTAAATTGTGTTACAATGGGTTTCGCTCAAGGCGCAAGGAGCACACACA |
| J1-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAAACCCATTGTAACACAATTTAAGAAAATTGTCAA |
| J2-F | ATAATTGACATTTTTTTAGTTTTGAGTTACAATGGTTGTCGCTCAAGGCGCAAGGAGCACACACA |
| J2-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGACAACCATTGTAACTCAAAACTAAAAAAATGTCAA |
| SP-PRO-F | ATAAGTTTACTCATATCCAGTCACAGAAACTGAACTATCTCGCTCAAGGCGCAAGGAGCACACACA |
| SP-TRO-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGATAGTTCAGTTTCTGTGACTGGATATGAGTAAAC |
| T7-CG-F | AATTCCGGCCGCGGGGCCCGCTTCGGCGGGCCCCGCGGCCGTTTTTTTAAACTGAACTATCG |
| T7-CG-R | GATCCGATAGTTCAGTTTAAAAAAACGGCCGCGGGGCCCGCCGAAGCGGGCCCCGCGGCCGG |
| WEAK-F | AATTCAGCCCCTCAGTATAGGGGCGTATTTTCTCAAACTGAACTATCG |
| WEAK-R | GATCCGATAGTTCAGTTTGAGAAAATACGCCCCTATACTGAGGGGCTG |
Table 2 Primers used in this study
| Primer | Description |
|---|---|
| pheA-F | TAGCAACAATAAGGCCTCCCAAATCGGGGGGCCTTTTTTATTGAT |
| pheA-R | TTAGATCAATAAAAAAGGCCCCCCGATTTGGGAGGCCTTATTGTT |
| thrL-F | TAGCTCAAAAAAGCCCGCACCTGACAGTGCGGGCTTTTTTTTTACT |
| thrL-R | TTAGAGTAAAAAAAAAGCCCGCACTGTCAGGTGCGGGCTTTTTTGA |
| rpsO-F | TAGCCAGAAAAGGGGGCCTGAGTGGCCCCTTTTTTCAAGCT |
| rpsO-R | TTAGAGCTTGAAAAAAGGGGCCACTCAGGCCCCCTTTTCTG |
| arcA-F | TAGCAATAAAAACGGCGCTAAAAAGCGCCGTTTTTTTTGACG |
| arcA-R | TTAGCGTCAAAAAAAACGGCGCTTTTTAGCGCCGTTTTTATT |
| fhuE-F | TAGCGTAAAAAAGGCAGCCATCTGGCTGCCTTAGTCTCCCCA |
| fhuE-R | TTAGTGGGGAGACTAAGGCAGCCAGATGGCTGCCTTTTTTAC |
| hisI-F | TAGCCCCTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGT |
| hisI-R | TTAGACGCGAAGTAATCTTTTCGGTTTTAAAGAAAAAGGGCAGGG |
| recA-F | TAGCAAGCAAAAGGGCCGCAGATGCGACCCTTGTGTATCAAC |
| recA-R | TTAGGTTGATACACAAGGGTCGCATCTGCGGCCCTTTTGCTT |
| T7-MOD-F | TAGCAAACAGATAGGCCCTCttcgGAGGGCCtatctgttTTTTTTT |
| T7-MOD-R | TTAGAAAAAAAaacagataGGCCCTCcgaaGAGGGCCTATCTGTTT |
| SP-TE-F | TAGCCCAATGTTTACTCATATCCAGTCACAGAAACTGAACTATC |
| SP-TE-R | TTAGGATAGTTCAGTTTCTGTGACTGGATATGAGTAAACATTGG |
| SPACE-5-F | CTAACTATC |
| SPACE-5-R | TTATGATAG |
| SPACE-13-F | CTAAAAACTGAACTATC |
| SPACE-13-R | TTATGATAGTTCAGTTT |
| SPACE-20-F | CTAAGTCACAGAAACTGAACTATC |
| SPACE-20-R | TTATGATAGTTCAGTTTCTGTGAC |
| SPACE-28-F | CTAACATATCCAGTCACAGAAACTGAACTATC |
| SPACE-28-R | TTATGATAGTTCAGTTTCTGTGACTGGATATG |
| SPACE-35-F | CTAAGTTTACTCATATCCAGTCACAGAAACTGAACTATC |
| SPACE-35-R | TTATGATAGTTCAGTTTCTGTGACTGGATATGAGTAAAC |
| SPACE-40-F | CTAACCAATGTTTACTCATATCCAGTCACAGAAACTGAACTATC |
| SPACE-40-R | TTATGATAGTTCAGTTTCTGTGACTGGATATGAGTAAACATTGG |
| dap-A16-F | ATAATTGTTTAACCCCCAAATGAGGGAAGAAGGTATAATTGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dap-A16-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCAATTATACCTTCTTCCCTCATTTGGGGGTTAAACAA |
| dap-e-F | ATAATTGTTTAGCCACCAAATGAGGGAAAGAGGCACAATGGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dap-e-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCCATTGTGCCTCTTTCCCTCATTTGGTGGCTAAACAA |
| dapA-e10-F | ATAATTGTTTTGACACCAAATGAGGGAATGTGGTATAATTGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dapA-e10-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCAATTATACCACATTCCCTCATTTGGTGTCAAAACAA |
| dapA-e11-F | ATAATTGTTTTGACACCAAATGAGGGAATGTGCTATAATGGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dapA-e11-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCCATTATAGCACATTCCCTCATTTGGTGTCAAAACAA |
| dapA-e12-F | ATAATTGTTTTGACACCAAATGAGGGAATGTGGTAGAGTGGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dapA-e12-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCCACTCTACCACATTCCCTCATTTGGTGTCAAAACAA |
| dapA-e10-35-F | ATAATTGTTTAACCCCCAAATGAGGGAATGTGGTATAATTGAACTCTCGCTCAAGGCGCAAGGAGCACACACA |
| dapA-e10-35-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGAGTTCAATTATACCACATTCCCTCATTTGGGGGTTAAACAA |
| J1-F | ATAATTGACAATTTTCTTAAATTGTGTTACAATGGGTTTCGCTCAAGGCGCAAGGAGCACACACA |
| J1-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAAACCCATTGTAACACAATTTAAGAAAATTGTCAA |
| J2-F | ATAATTGACATTTTTTTAGTTTTGAGTTACAATGGTTGTCGCTCAAGGCGCAAGGAGCACACACA |
| J2-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGACAACCATTGTAACTCAAAACTAAAAAAATGTCAA |
| SP-PRO-F | ATAAGTTTACTCATATCCAGTCACAGAAACTGAACTATCTCGCTCAAGGCGCAAGGAGCACACACA |
| SP-TRO-R | TCATTGTGTGTGCTCCTTGCGCCTTGAGCGAGATAGTTCAGTTTCTGTGACTGGATATGAGTAAAC |
| T7-CG-F | AATTCCGGCCGCGGGGCCCGCTTCGGCGGGCCCCGCGGCCGTTTTTTTAAACTGAACTATCG |
| T7-CG-R | GATCCGATAGTTCAGTTTAAAAAAACGGCCGCGGGGCCCGCCGAAGCGGGCCCCGCGGCCGG |
| WEAK-F | AATTCAGCCCCTCAGTATAGGGGCGTATTTTCTCAAACTGAACTATCG |
| WEAK-R | GATCCGATAGTTCAGTTTGAGAAAATACGCCCCTATACTGAGGGGCTG |
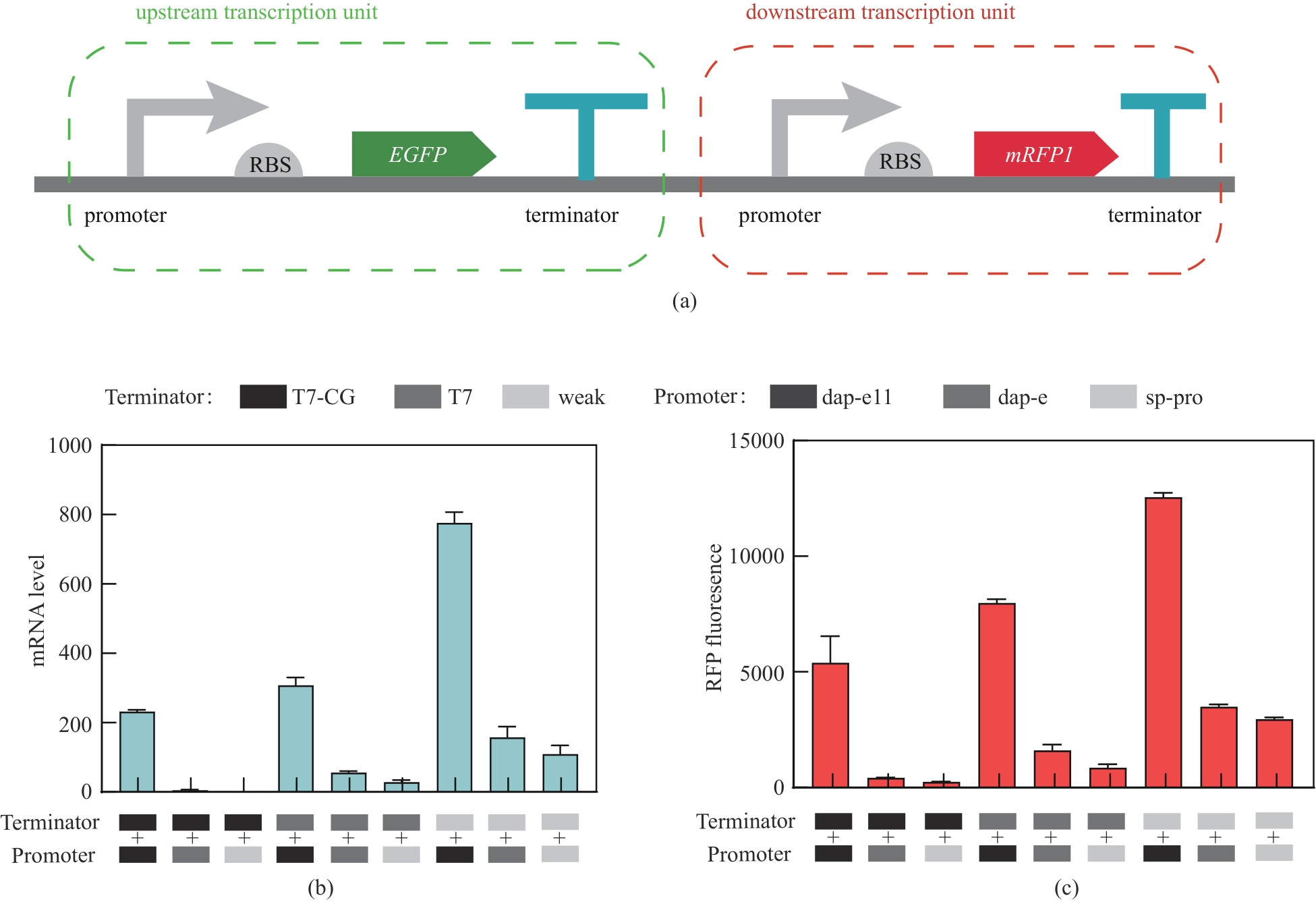
Fig. 1 Analysis of the regulatory effect of the combination of terminators and promoters with different strengthsSchematic structure of upstream and downstream transcriptional units in the plasmid (a), red fluorescent transcript levels (b), and expression levels (c) of downstream transcriptional units in the nine mutant plasmids
| Assembly | Part Name | Activity | Sequence(5′→3′) |
|---|---|---|---|
| Terminator | pheA | Strong ↓ weak | AATCATAAGGCCTGACAAATCGGGGGGGATTTTTTCTTGA |
| thrL | TCCAAAAAGCCCCGACCTGACAGTGACGGCTTTTTTATTAC | ||
| T7-MOD | AAACAGATAGGCCCTCTTCGGAGGGCCTATCTGTTTTTTTTT | ||
| arcA | AATATAAACGGCGCTAAAACGCGCGGTTTTTTTTGCC | ||
| rpsO | CAGCAAAGGGTGCCTGAGTGCTCCCTTTTTGCAAGC | ||
| fhuE | GTAAAAAAGGCAGCCATCTGGCTGCCTTAGTCTCCCCA | ||
| hisI | CCCTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGT | ||
| recA | AAGCAAAAGGGCCGCAGATGCGACCCTTGTGTATCAAC | ||
| SP-TE | CCAATGTTTACTCATATCCAGTCACAGAAACTGAACTATC | ||
| Spacer | SP-13 | Short ↓ Long | AAACTGAACTATC |
| SP-20 | GTCACAGAAACTGAACTATC | ||
| SP-28 | CATATCCAGTCACAGAAACTGAACTATC | ||
| SP-35 | GTTTACTCATATCCAGTCACAGAAACTGAACTATC | ||
| SP-40 | CCAATGTTTACTCATATCCAGTCACAGAAACTGAACTATC | ||
| Promoter | dapA-e12 | Strong ↓ weak | TTGTTTTGACACCAAATGAGGGAATGTGGTAGAGTGGAACTC |
| dapA-e11 | TTGTTTTGACACCAAATGAGGGAATGTGCTATAATGGAACTC | ||
| dapA-e10 | TTGTTTTGACACCAAATGAGGGAATGTGGTATAATTGAACTC | ||
| dap-A16 | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGTATAATTGAACTC | ||
| J2 | TTGACATTTTTTTAGTTTTGAGTTACAATGGTTG | ||
| dapA-e10-35 | TTGTTTAACCCCCAAATGAGGGAATGTGGTATAATTGAACTC | ||
| J1 | TTGACAATTTTCTTAAATTGTGTTACAATGGGTT | ||
| dap-e | TTGTTTAGCCACCAAATGAGGGAAAGAGGCACAATGGAACTC | ||
| SP-PRO | GTTTACTCATATCCAGTCACAGAAACTGAAC |
Table 3 Parts in the library
| Assembly | Part Name | Activity | Sequence(5′→3′) |
|---|---|---|---|
| Terminator | pheA | Strong ↓ weak | AATCATAAGGCCTGACAAATCGGGGGGGATTTTTTCTTGA |
| thrL | TCCAAAAAGCCCCGACCTGACAGTGACGGCTTTTTTATTAC | ||
| T7-MOD | AAACAGATAGGCCCTCTTCGGAGGGCCTATCTGTTTTTTTTT | ||
| arcA | AATATAAACGGCGCTAAAACGCGCGGTTTTTTTTGCC | ||
| rpsO | CAGCAAAGGGTGCCTGAGTGCTCCCTTTTTGCAAGC | ||
| fhuE | GTAAAAAAGGCAGCCATCTGGCTGCCTTAGTCTCCCCA | ||
| hisI | CCCTGCCCTTTTTCTTTAAAACCGAAAAGATTACTTCGCGT | ||
| recA | AAGCAAAAGGGCCGCAGATGCGACCCTTGTGTATCAAC | ||
| SP-TE | CCAATGTTTACTCATATCCAGTCACAGAAACTGAACTATC | ||
| Spacer | SP-13 | Short ↓ Long | AAACTGAACTATC |
| SP-20 | GTCACAGAAACTGAACTATC | ||
| SP-28 | CATATCCAGTCACAGAAACTGAACTATC | ||
| SP-35 | GTTTACTCATATCCAGTCACAGAAACTGAACTATC | ||
| SP-40 | CCAATGTTTACTCATATCCAGTCACAGAAACTGAACTATC | ||
| Promoter | dapA-e12 | Strong ↓ weak | TTGTTTTGACACCAAATGAGGGAATGTGGTAGAGTGGAACTC |
| dapA-e11 | TTGTTTTGACACCAAATGAGGGAATGTGCTATAATGGAACTC | ||
| dapA-e10 | TTGTTTTGACACCAAATGAGGGAATGTGGTATAATTGAACTC | ||
| dap-A16 | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGTATAATTGAACTC | ||
| J2 | TTGACATTTTTTTAGTTTTGAGTTACAATGGTTG | ||
| dapA-e10-35 | TTGTTTAACCCCCAAATGAGGGAATGTGGTATAATTGAACTC | ||
| J1 | TTGACAATTTTCTTAAATTGTGTTACAATGGGTT | ||
| dap-e | TTGTTTAGCCACCAAATGAGGGAAAGAGGCACAATGGAACTC | ||
| SP-PRO | GTTTACTCATATCCAGTCACAGAAACTGAAC |
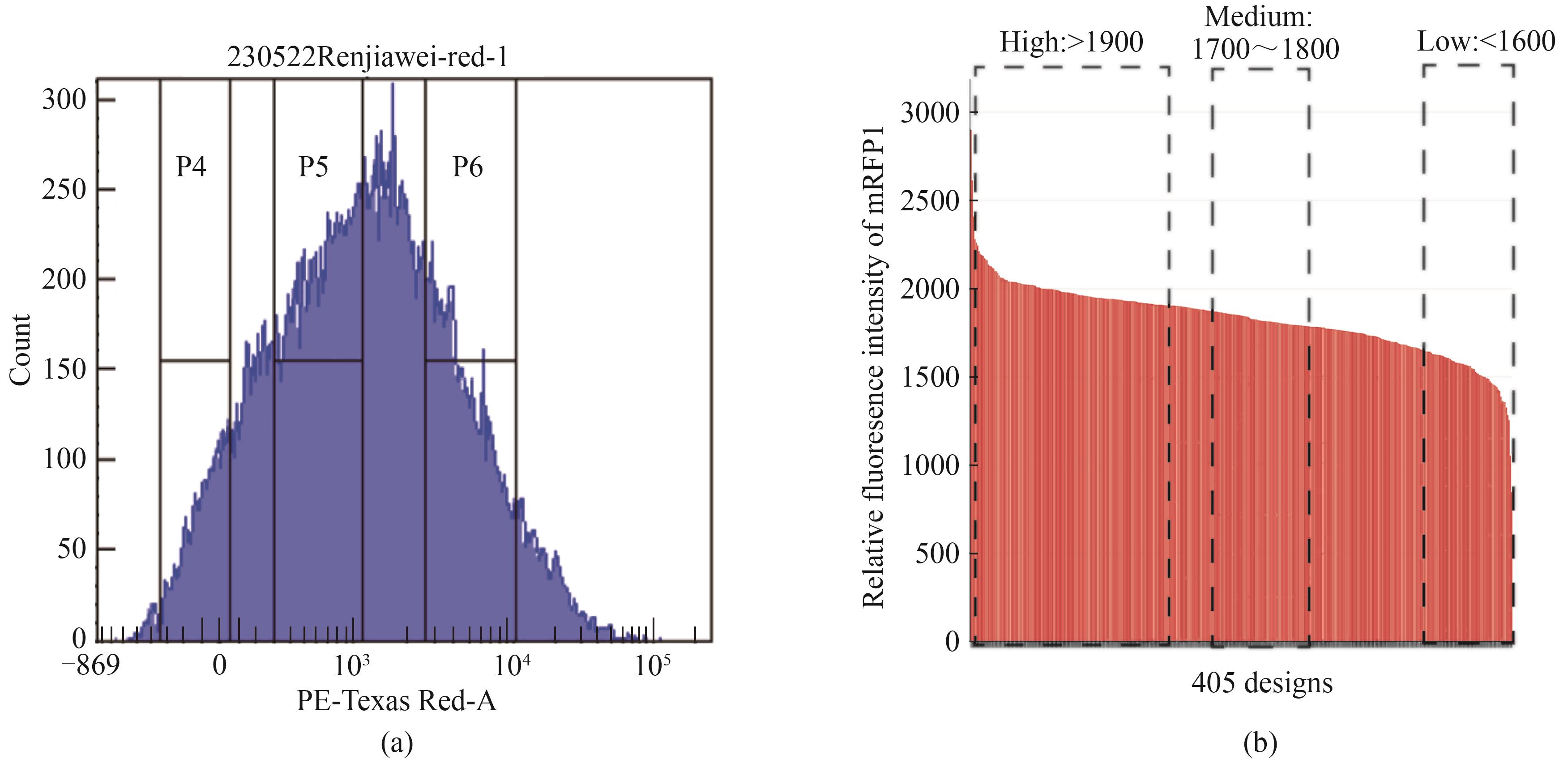
Fig. 2 Expression of downstream genes regulated by the assemblies of terminator-spacer-promoter(a) Screening of combinatorial elements by flow cytometry based on the red fluorescence for the expression of targeted genes; (b) Grouping of the assemblies of terminator-spacer-promoter into High, Medium, and Low expression groups based on the relative fluorescence intensity of mRFP1 associated with the expression of targeted genes
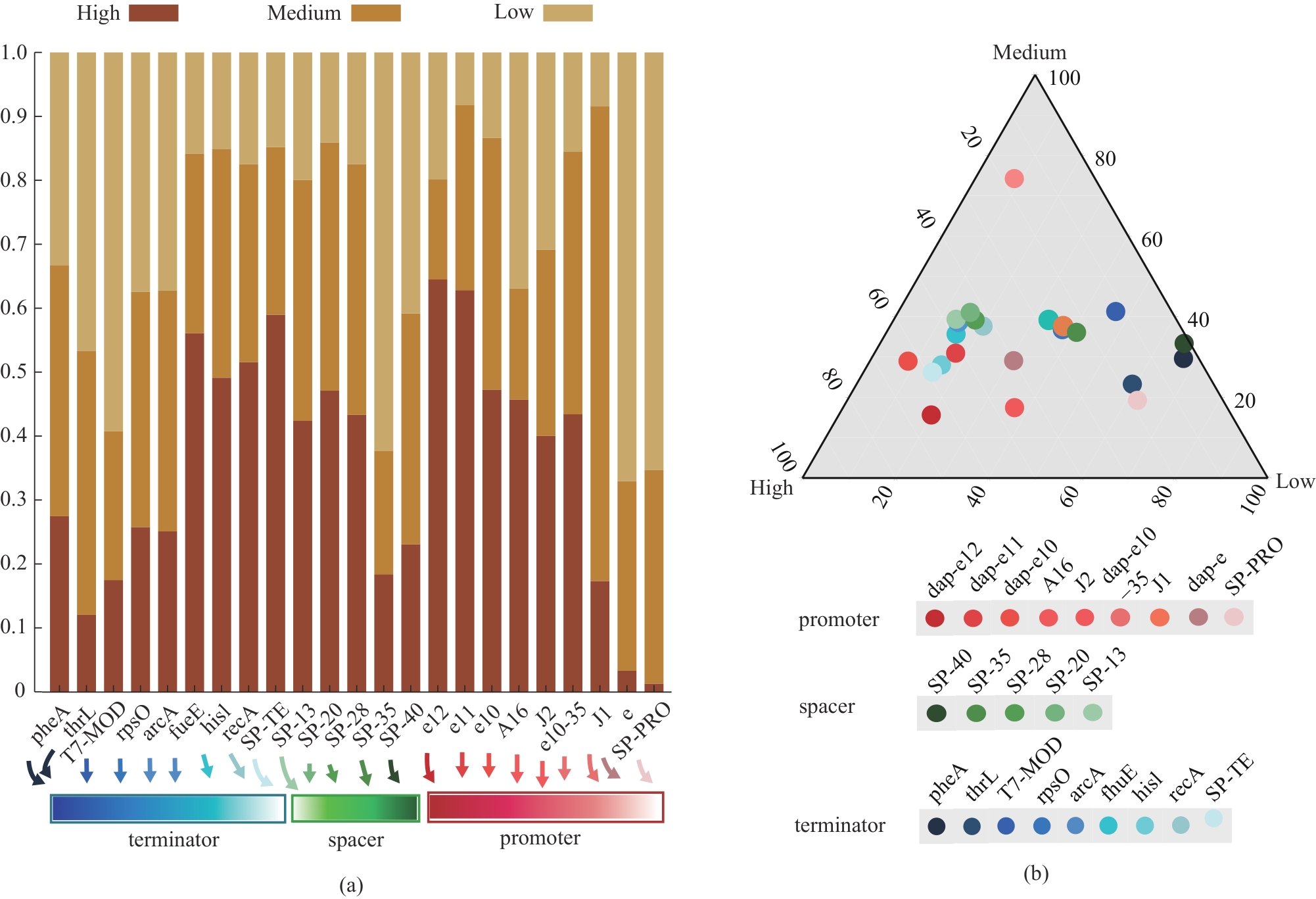
Fig. 3 Frequency of each part used in the assemblies at the “High”, “Medium”, and “Low” groups(a) Stacked histograms of the occurrence frequency of different components in the “High”, “Medium”, and “Low” groups; (b) Ternary results of the occurrence frequency of different components in the “High”, “Medium”, and “Low” groups
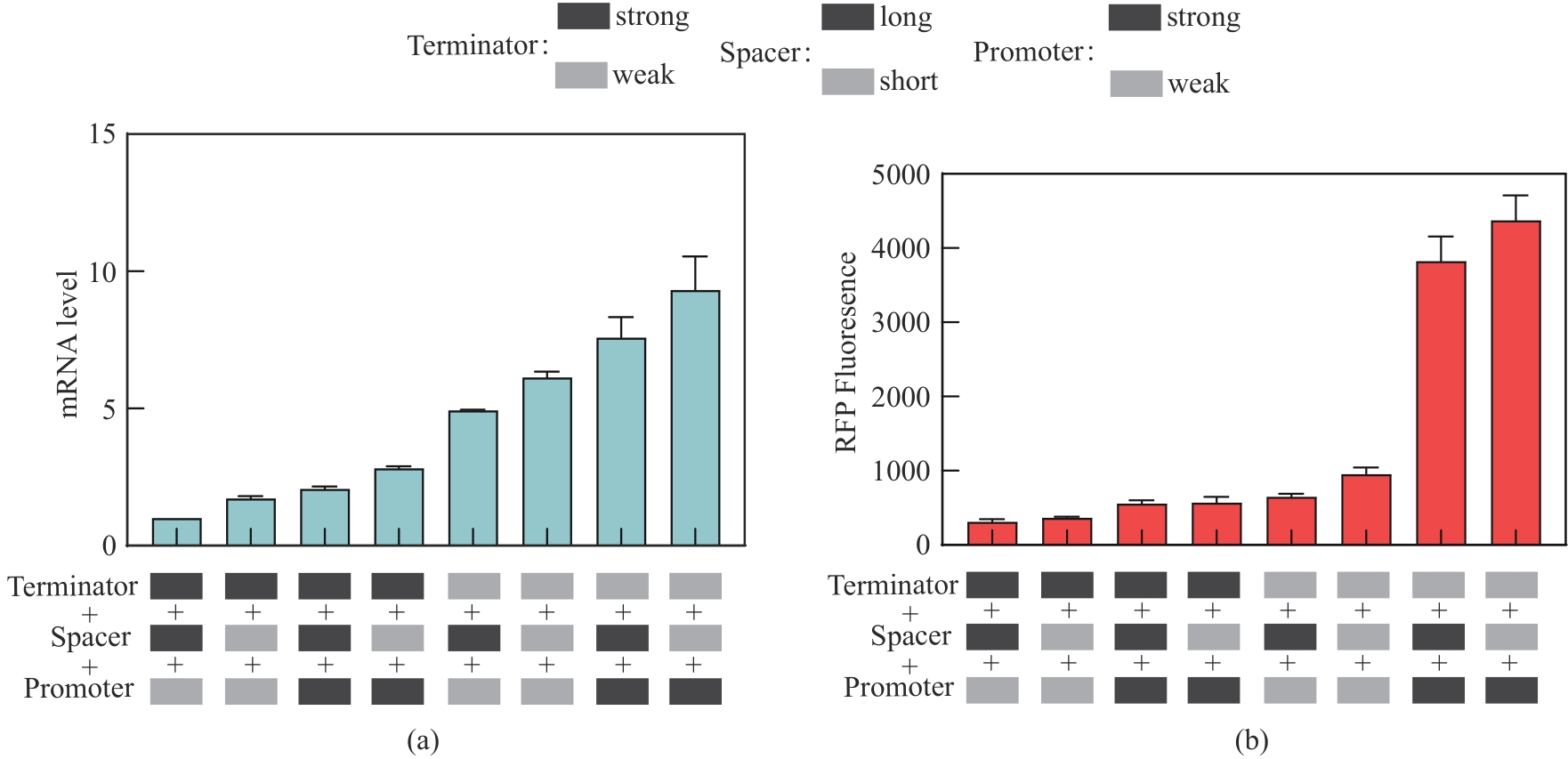
Fig. 4 Expression levels of mRNA and red fluorescent protein with the downstream transcription unit regulated by typical assemblies selected from the one-pot assembly library(The strong and weak promoters were dap-e11 and dap-e, the strong and weak terminators were pheA and recA, and the long and short interval sequences were sp-40 and sp-13, respectively.)
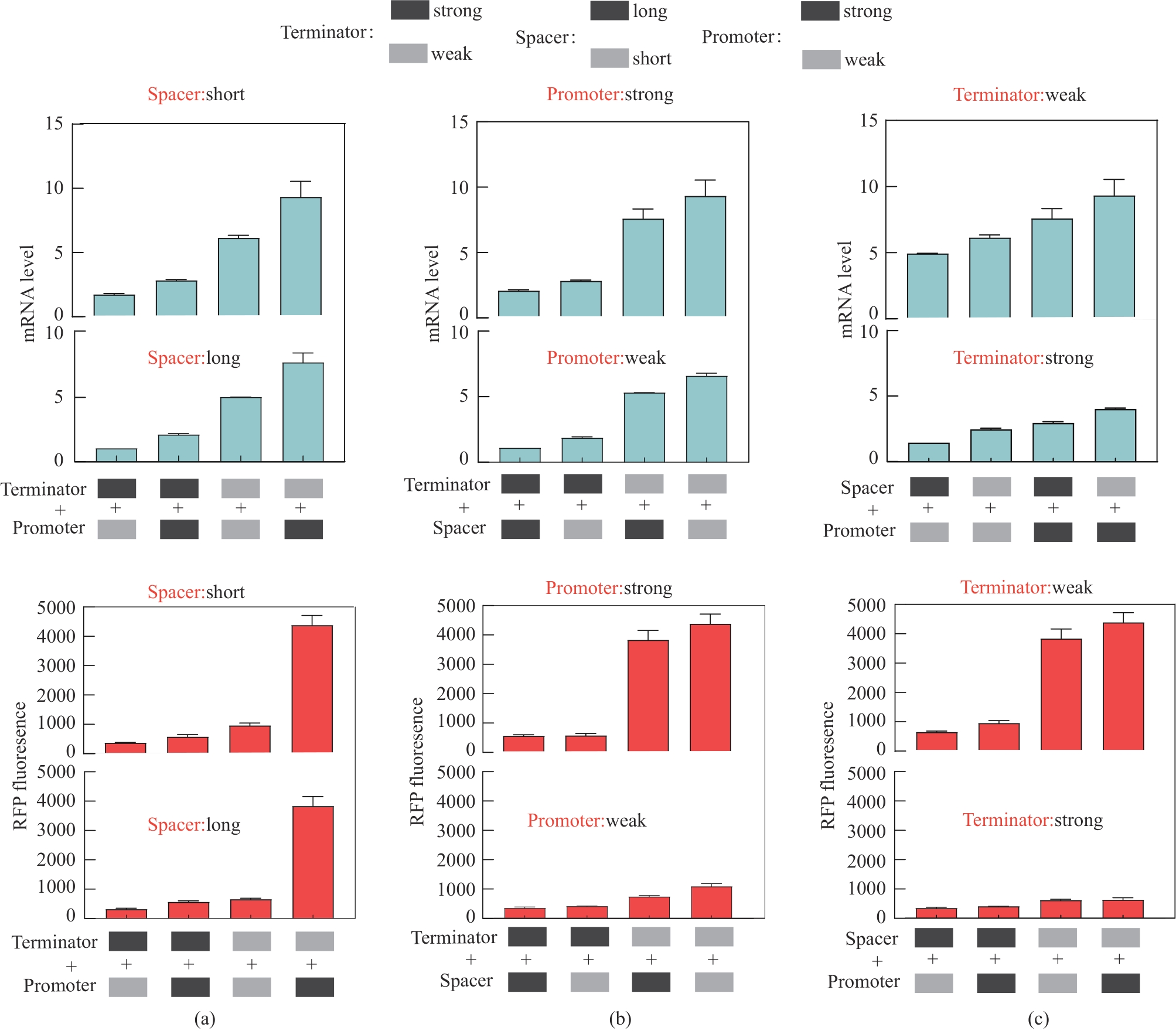
Fig. 5 Influence of the combination of promoters and terminators on expression of the mRNA and fluorescence protein(a) Effect of different terminator and promoter combinations on gene expression with fixed interval sequences; (b) Effect of different spacer sequences and terminator combinations on gene expression with fixed promoters; (c) Effects of different promoters and combinations of promoters on gene expression with fixed terminators. The strong and weak promoters were dap-e11 and dap-e, the strong and weak terminators were pheA and recA, and the long and short interval sequences were sp-40 and sp-13, respectively.
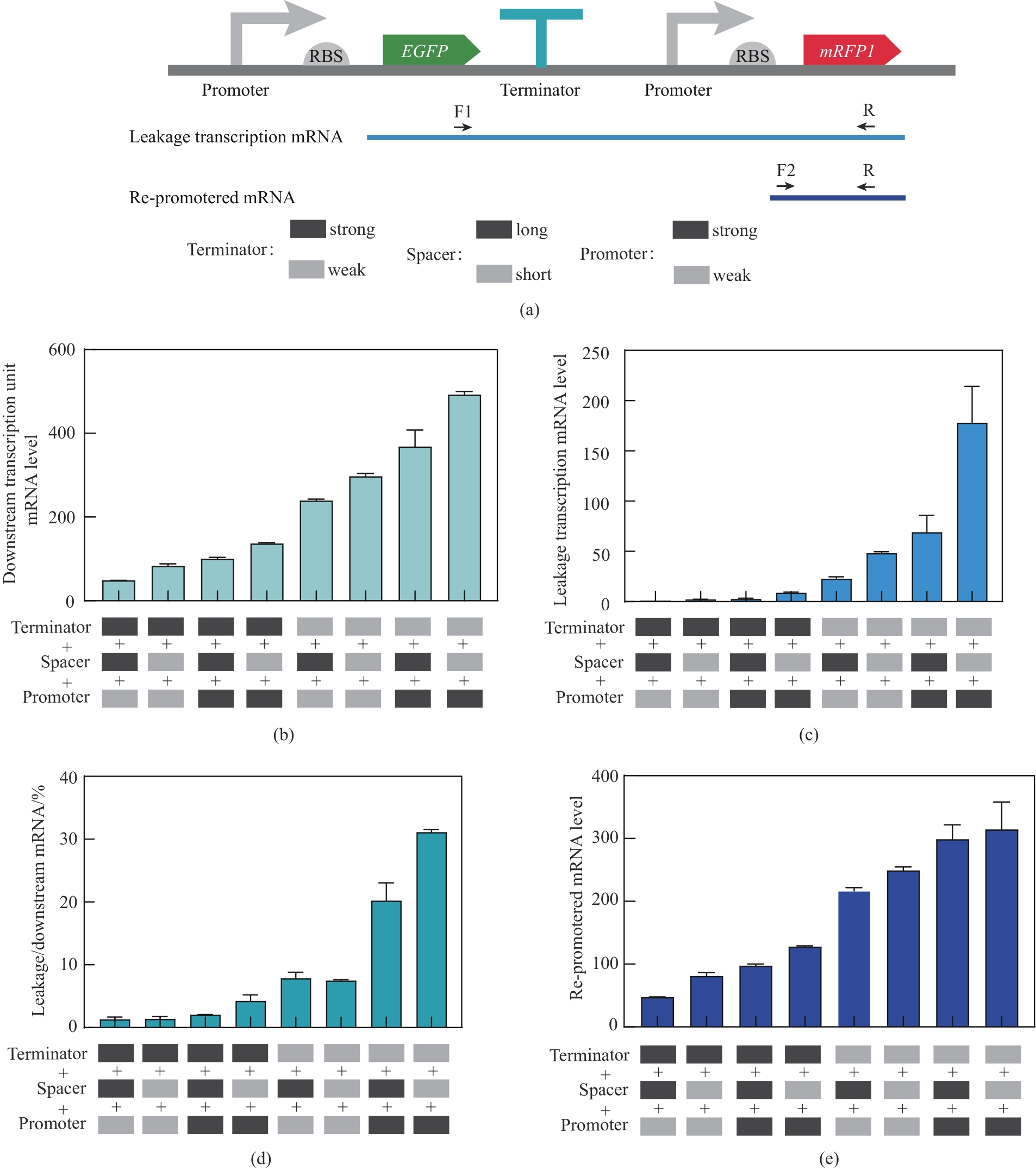
Fig. 6 Schematic representation of downstream mRNA, leakage transcription mRNA, and re-promoted mRNA levels under the regulation of different combinatorial elements.(a) Schematic representation of leakage transcription mRNA and re-promoted mRNA; (b) Downstream mRNA levels under the regulation of different combinatorial elements; (c) Leakage transcription mRNA levels under the regulation of different combinatorial elements; (d) Percentage of Leakage transcription mRNA in downstream mRNA; (e) Re-promoted mRNA levels under the regulation of different combinatorial elements; The strong and weak promoters were dap-e11 and dap-e, the strong and weak terminators were pheA and recA, and the long and short interval sequences were sp-40 and sp-13, respectively.
| 1 | LAURSEN B S, SØRENSEN H P, MORTENSEN K K, et al. Initiation of protein synthesis in bacteria[J]. Microbiology and Molecular Biology Reviews, 2005, 69(1): 101-123. |
| 2 | MORENO J M, SØRENSEN H P, MORTENSEN K K, et al. Macromolecular mimicry in translation initiation: a model for the initiation factor IF2 on the ribosome[J]. IUBMB Life, 2000, 50(6): 347-354. |
| 3 | RAMAKRISHNAN V. Ribosome structure and the mechanism of translation[J]. Cell, 2002, 108(4): 557-572. |
| 4 | ZHOU S H, DU G C, KANG Z, et al. The application of powerful promoters to enhance gene expression in industrial microorganisms[J]. World Journal of Microbiology & Biotechnology, 2017, 33(2): 23. |
| 5 | XU N, WEI L, LIU J. Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis[J]. World Journal of Microbiology & Biotechnology, 2019, 35(2): 33. |
| 6 | ZALATAN F, PLATT T. Effects of decreased cytosine content on rho interaction with the rho-dependent Terminator trp t′ in Escherichia coli [J]. Journal of Biological Chemistry, 1992, 267(27): 19082-19088. |
| 7 | HENKIN T M, YANOFSKY C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions[J]. BioEssays, 2002, 24(8): 700-707. |
| 8 | ABE H, ABO T, AIBA H. Regulation of intrinsic Terminator by translation in Escherichia coli: transcription termination at a distance downstream[J]. Genes to Cells, 1999, 4(2): 87-97. |
| 9 | RAY-SONI A, BELLECOURT M J, LANDICK R. Mechanisms of bacterial transcription termination: all good things must end[J]. Annual Review of Biochemistry, 2016, 85: 319-347. |
| 10 | LEE D N, PHUNG L, STEWART J, et al. Transcription pausing by Escherichia coli RNA polymerase is modulated by downstream DNA sequences[J]. Journal of Biological Chemistry, 1990, 265(25): 15145-15153. |
| 11 | KIREEVA M L, KASHLEV M. Mechanism of sequence-specific pausing of bacterial RNA polymerase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(22): 8900-8905. |
| 12 | CHAN C L, WANG D G, LANDICK R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3΄ end spacing distinguishes pause and termination pathways [J]. Journal of Molecular Biology, 1997, 268(1): 54-68. |
| 13 | MAIRHOFER J, WITTWER A, CSERJAN-PUSCHMANN M, et al. Preventing T7 RNA polymerase read-through transcription-a synthetic termination signal capable of improving bioprocess stability[J]. ACS Synthetic Biology, 2015, 4(3): 265-273. |
| 14 | ZHANG B, YU M, ZHOU Y, et al. Improvement of L-ornithine production by attenuation of argF in engineered Corynebacterium glutamicum S9114[J]. AMB Express, 2018, 8(1): 26. |
| 15 | JU X W, LI D Y, LIU S X. Full-length RNA profiling reveals pervasive bidirectional transcription terminators in bacteria[J]. Nature Microbiology, 2019, 4(11): 1907-1918. |
| 16 | AHN J H, KANG T J, KIM D M. Tuning the expression level of recombinant proteins by modulating mRNA stability in a cell-free protein synthesis system[J]. Biotechnology and Bioengineering, 2008, 101(2): 422-427. |
| 17 | CHEN Y J, LIU P, NIELSEN A A K, et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints[J]. Nature Methods, 2013, 10(7): 659-664. |
| 18 | CUI W J, LIN Q, HU R C, et al. Data-driven and in silico-assisted design of broad host-range minimal intrinsic terminators adapted for bacteria[J]. ACS Synthetic Biology, 2021, 10(6): 1438-1450. |
| 19 | ZHAI W J, DUAN Y T, ZHANG X M, et al. Sequence and thermodynamic characteristics of terminators revealed by FlowSeq and the discrimination of terminators strength[J]. Synthetic and Systems Biotechnology, 2022, 7(4): 1046-1055. |
| 20 | 翟伟绩. 大肠杆菌终止子文库构建及构效关系分析[D]. 无锡: 江南大学, 2022. |
| ZHAI W J. The construction of the terminator library and analysis of the structure-activity relationship[D]. Wuxi: Jiangnan University, 2022. | |
| 21 | DU L P, GAO R, FORSTER A C. Engineering multigene expression in vitro and in vivo with small terminators for T7 RNA polymerase[J]. Biotechnology and Bioengineering, 2009, 104(6): 1189-1196. |
| 22 | TARNOWSKI M J, GOROCHOWSKI T E. Massively parallel characterization of engineered transcript isoforms using direct RNA sequencing[J]. Nature Communications, 2022, 13(1): 434. |
| 23 | LI R, ZHANG Q, LI J B, et al. Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the rho-independent terminator[J]. Nucleic Acids Research, 2016, 44(6): 2554-2563. |
| 24 | LALANNE J B, TAGGART J C, GUO M S, et al. Evolutionary convergence of pathway-specific enzyme expression stoichiometry[J]. Cell, 2018, 173(3): 749-761.e38. |
| 25 | GOROCHOWSKI T E, ESPAH BORUJENI A, PARK Y, et al. Genetic circuit characterization and debugging using RNA-seq[J]. Molecular Systems Biology, 2017, 13(11): 952. |
| 26 | KOSURI S, GOODMAN D B, CAMBRAY G, et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(34): 14024-14029. |
| 27 | CAMBRAY G, GUIMARAES J C, ARKIN A P. Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli [J]. Nature Biotechnology, 2018, 36: 1005-1015. |
| 28 | EVFRATOV S A, OSTERMAN I A, KOMAROVA E S, et al. Application of sorting and next generation sequencing to study 5′-UTR influence on translation efficiency in Escherichia coli [J]. Nucleic Acids Research, 2017, 45(6): 3487-3502. |
| 29 | YOON B J. Hidden Markov models and their applications in biological sequence analysis[J]. Current Genomics, 2009, 10(6): 402-415. |
| 30 | KUO S T, JAHN R L, CHENG Y J, et al. Global fitness landscapes of the Shine-Dalgarno sequence[J]. Genome Research, 2020, 30(5): 711-723. |
| 31 | OSTERMAN I A, CHERVONTSEVA Z S, EVFRATOV S A, et al. Translation at first sight: the influence of leading codons[J]. Nucleic Acids Research, 2020, 48(12): 6931-6942. |
| 32 | VERMA M, CHOI J, COTTRELL K A, et al. A short translational ramp determines the efficiency of protein synthesis[J]. Nature Communications, 2019, 10(1): 5774. |
| 33 | GAO R, YU K, NIE J K, et al. Deep sequencing reveals global patterns of mRNA recruitment during translation initiation[J]. Scientific Reports, 2016, 6: 30170. |
| 34 | LESNIK E A, SAMPATH R, LEVENE H B, et al. Prediction of rho-independent transcriptional terminators in Escherichia coli [J]. Nucleic Acids Research, 2001, 29(17): 3583-3594. |
| 35 | DUAN Y T, ZHAI W J, LIU W J, et al. Fine-tuning multi-gene clusters via well-characterized gene expression regulatory elements: case study of the arginine synthesis pathway in C. glutamicum [J]. ACS Synthetic Biology, 2021, 10(1): 38-48. |
| 36 | HUDSON A J, WIEDEN H J. Rapid generation of sequence-diverse terminator libraries and their parameterization using quantitative Term-Seq[J]. Synthetic Biology, 2019, 4(1): ysz026. |
| [1] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| [2] | Huimin YU, Yukun ZHENG, Yan DU, Miaomiao WANG, Youxiang LIANG. Microbial promoter engineering strategies in synthetic biology [J]. Synthetic Biology Journal, 2021, 2(4): 598-611. |
| [3] | Yue SHENG, Genlin ZHANG. Yeast terminator engineering: from mechanism exploration to artificial design [J]. Synthetic Biology Journal, 2020, 1(6): 709-721. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||