Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (5): 1072-1092.DOI: 10.12211/2096-8280.2025-089
• Invited Review • Previous Articles Next Articles
Plant artificial chromosomes: current research progress and future application perspectives
PU Ya1,2, JIAO Yuling1,2
- 1.State Key Laboratory of Gene Function and Modulation Research,School of Life Sciences,Peking University,Beijing 100871,China
2.Peking -Tsinghua Center for Life Sciences,Center for Quantitative Biology,Academy for Advanced Interdisciplinary Studies,Peking University,Beijing 100871,China
-
Received:2025-09-01Revised:2025-09-26Online:2025-11-05Published:2025-10-31 -
Contact:JIAO Yuling
植物人工染色体的研究现状与应用前景
蒲娅1,2, 焦雨铃1,2
- 1.北京大学生命科学学院,基因功能研究与操控全国重点实验室,北京 100871
2.北京大学前沿交叉学科研究院,北京大学-清华大学生命科学联合中心,北京大学定量生物学中心,北京 100871
-
通讯作者:焦雨铃 -
作者简介:蒲娅 (1994—),女,博士后。研究方向为植物人工着丝粒的构建与功能研究。E-mail:puya0824@pku.edu.cn焦雨铃 (1979—),男,教授,博士生导师。研究方向为植物发育生物学与合成生物学等。E-mail:yuling.jiao@pku.edu.cn -
基金资助:国家重点研发计划(2023YFE0101100);国家重点研发计划(2024YFF1000704)
CLC Number:
Cite this article
PU Ya, JIAO Yuling. Plant artificial chromosomes: current research progress and future application perspectives[J]. Synthetic Biology Journal, 2025, 6(5): 1072-1092.
蒲娅, 焦雨铃. 植物人工染色体的研究现状与应用前景[J]. 合成生物学, 2025, 6(5): 1072-1092.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-089
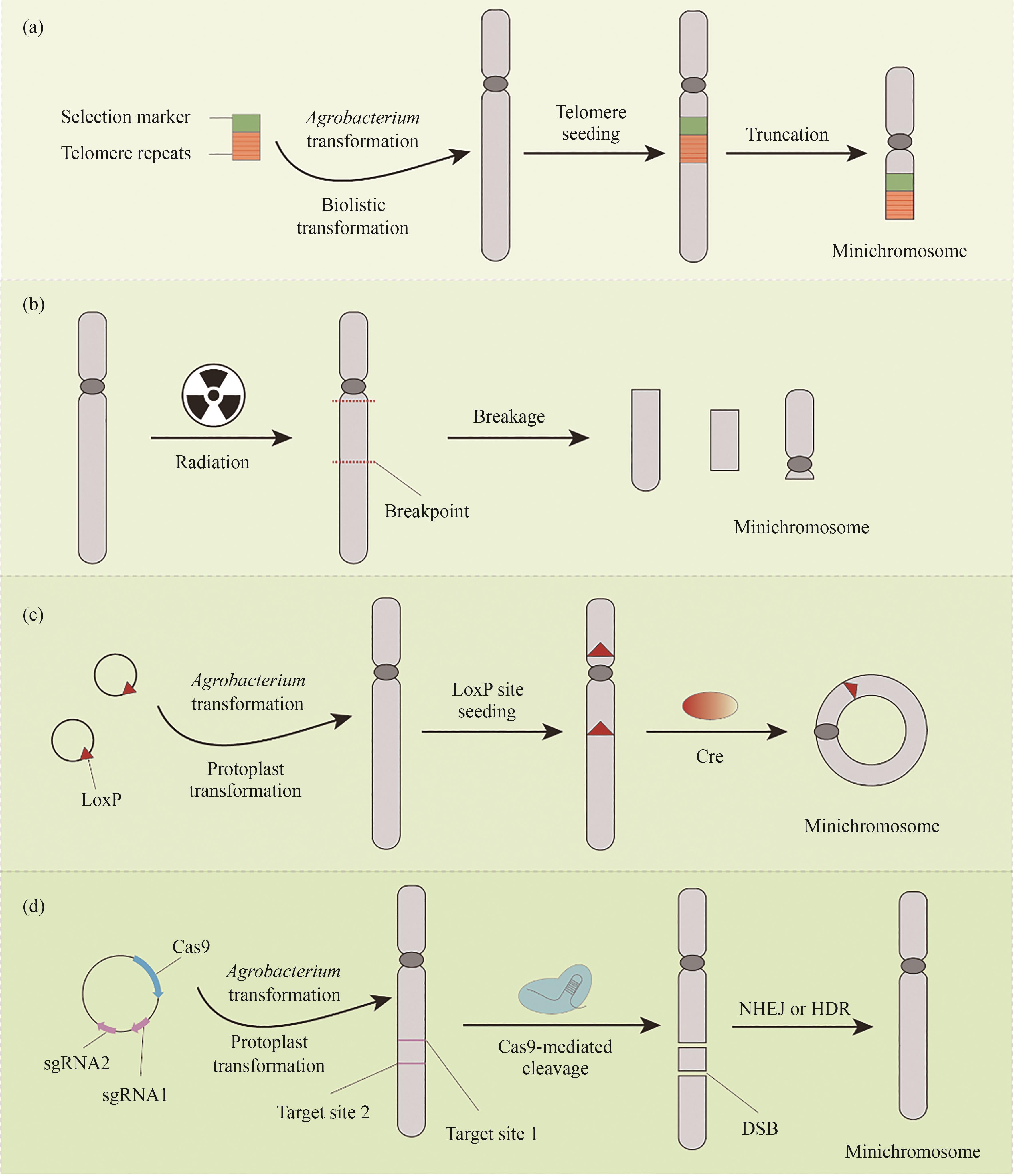
Fig. 1 Top-down approach for minichromosome construction in plant(a) Telomere-mediated chromosome truncation; (b) Ionizing radiation-induced chromosome breakage; (c) Site-specific recombinase-mediated chromosome truncation; (d) CRISPR/Cas9-mediated chromosome deletionNHEJ—the non-homologous DNA end joining; HDR—the homology-directed recombination
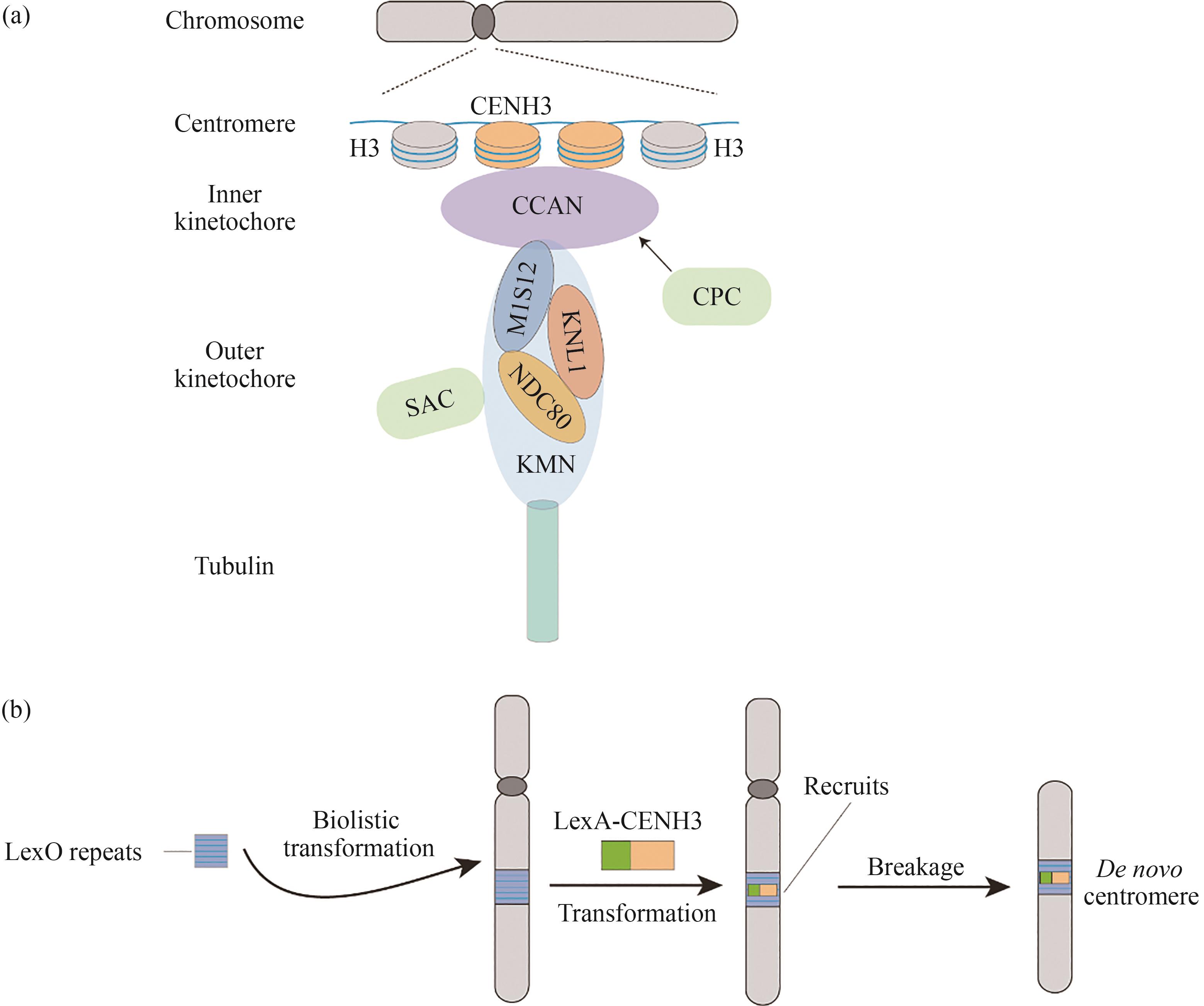
Fig. 2 An organization of the kinetochore and the synthesis strategy of the centromere(a) Centromere is defined by specific CENH3 nucleosomes, which are responsible for recruiting kinetochore proteins. The kinetochore comprises an inner constitutive centromere-associated network (CCAN) and an outer KMN network (KNL1-MIS12-NDC80). Kinetochore assembly and its attachment to microtubule also involve kinetochore regulators, such as spindle assembly checkpoint (SAC) and chromosome passenger complex (CPC). (b) De novo centromere formation is achieved by recruiting LexA-CENH3 to the LexO repeat array inserted into the genome.
转化方法 Transformation method | 物种 Species | 靶向组织 Targeted tissue | 递送的最大片段 The maximum size of the delivered fragment | 局限性 Limitations |
|---|---|---|---|---|
| 农杆菌介导转化 | 水稻[ | 种子来源的愈伤组织 | 164 kb | 宿主限制,片段随机插入,周期较长 |
| 基因枪粒子递送 | 烟草[ | 悬浮细胞 | 150 kb | 大片段易断裂,转化效率较低,设备昂贵 |
| 纳米材料递送 | 棉花[ | 花粉 | 15 kb | 普适性待定,递送效率较低 |
| PEG介导转化 | 烟草[ | 原生质体(叶片来源) | 300 kb | 细胞毒性,单细胞再生效率低 |
| 电穿孔法 | 灵芝[ | 原生质体(菌丝来源) | 100 kb | 大片段易断裂,组织损伤 |
Table 1 Summary of different delivery methods of large DNA fragments in plants
转化方法 Transformation method | 物种 Species | 靶向组织 Targeted tissue | 递送的最大片段 The maximum size of the delivered fragment | 局限性 Limitations |
|---|---|---|---|---|
| 农杆菌介导转化 | 水稻[ | 种子来源的愈伤组织 | 164 kb | 宿主限制,片段随机插入,周期较长 |
| 基因枪粒子递送 | 烟草[ | 悬浮细胞 | 150 kb | 大片段易断裂,转化效率较低,设备昂贵 |
| 纳米材料递送 | 棉花[ | 花粉 | 15 kb | 普适性待定,递送效率较低 |
| PEG介导转化 | 烟草[ | 原生质体(叶片来源) | 300 kb | 细胞毒性,单细胞再生效率低 |
| 电穿孔法 | 灵芝[ | 原生质体(菌丝来源) | 100 kb | 大片段易断裂,组织损伤 |
| [1] | DAWE R K. Charting the path to fully synthetic plant chromosomes[J]. Experimental Cell Research, 2020, 390(1): 111951. |
| [2] | KAN M M, HUANG T B, ZHAO P P. Artificial chromosome technology and its potential application in plants[J]. Frontiers in Plant Science, 2022, 13: 970943. |
| [3] | BIRCHLER J A, KELLY J, SINGH J, et al. Synthetic minichromosomes in plants: past, present, and promise[J]. The Plant Journal, 2024, 120(6): 2356-2366. |
| [4] | MURRAY A W, SZOSTAK J W. Construction of artificial chromosomes in yeast[J]. Nature, 1983, 305(5931): 189-193. |
| [5] | FARR C, FANTES J, GOODFELLOW P, et al. Functional reintroduction of human telomeres into mammalian cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 1991, 88(16): 7006-7010. |
| [6] | SHIZUYA H, BIRREN B, KIM U J, et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector[J]. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(18): 8794-8797. |
| [7] | HAMILTON C M, FRARY A, LEWIS C, et al. Stable transfer of intact high molecular weight DNA into plant chromosomes[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(18): 9975-9979. |
| [8] | LIU Y G, SHIRANO Y, FUKAKI H, et al. Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(11): 6535-6540. |
| [9] | PUCHTA H, HOUBEN A. Plant chromosome engineering-past, present and future[J]. New Phytologist, 2024, 241(2): 541-552. |
| [10] | BIRCHLER J A. Engineered minichromosomes in plants[J]. Chromosome Research, 2015, 23(1): 77-85. |
| [11] | CHOO K H A. Engineering human chromosomes for gene therapy studies[J]. Trends in Molecular Medicine, 2001, 7(6): 235-237. |
| [12] | SAFFERY R, WONG L H, IRVINE D V, et al. Construction of neocentromere-based human minichromosomes by telomere-associated chromosomal truncation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(10): 5705-5710. |
| [13] | HELLER R, BROWN K E, BURGTORF C, et al. Mini-chromosomes derived from the human Y chromosome by telomere directed chromosome breakage[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(14): 7125-7130. |
| [14] | Analysis of meiotic chromosome pairing in the female mouse using a novel minichromosome[J]. Chromosome Research, 1998, 6(4): 269-276. |
| [15] | AURICHE C, DONINI P, ASCENZIONI F. Molecular and cytological analysis of a 5.5 Mb minichromosome[J]. EMBO Reports, 2001, 2(2): 102-107. |
| [16] | MORALLI D, VAGNARELLI P, BENSI M, et al. Insertion of aloxP site in a size-reduced human accessory chromosome[J]. Cytogenetics and Cell Genetics, 2001, 94(3-4): 113-120. |
| [17] | CARINE K, JACQUEMIN-SABLON A, WALTZER E, et al. Molecular characterization of human minichromosomes with centromere from chromosome 1 in human-hamster hybrid cells[J]. Somatic Cell and Molecular Genetics, 1989, 15(5): 445-460. |
| [18] | CSONKA E, CSERPÁN I, FODOR K, et al. Novel generation of human satellite DNA-based artificial chromosomes in mammalian cells[J]. Journal of Cell Science, 2000, 113(Pt 18): 3207-3216. |
| [19] | LAZAR N H, CELIK S, CHEN L, et al. High-resolution genome-wide mapping of chromosome-arm-scale truncations induced by CRISPR-Cas9 editing[J]. Nature Genetics, 2024, 56(7): 1482-1493. |
| [20] | UNO N, HIRAMATSU K, UNO K, et al. CRISPR/Cas9-induced transgene insertion and telomere-associated truncation of a single human chromosome for chromosome engineering in CHO and A9 cells[J]. Scientific Reports, 2017, 7: 12739. |
| [21] | NAHMAD A D, REUVENI E, GOLDSCHMIDT E, et al. Frequent aneuploidy in primary human T cells after CRISPR-Cas9 cleavage[J]. Nature Biotechnology, 2022, 40(12): 1807-1813. |
| [22] | FAJKUS J, SÝKOROVÁ E, LEITCH A R. Techniques in plant telomere biology[J]. BioTechniques, 2005, 38(2): 233-243. |
| [23] | YU W C, LAMB J C, HAN F P, et al. Telomere-mediated chromosomal truncation in maize[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(46): 17331-17336. |
| [24] | YU W C, HAN F P, GAO Z, et al. Construction and behavior of engineered minichromosomes in maize[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(21): 8924-8929. |
| [25] | BIRCHLER J A, YANG H. The supernumerary B chromosome of maize: drive and genomic conflict[J]. Open Biology, 2021, 11(11): 210197. |
| [26] | CHEN J Y, BIRCHLER J A, HOUBEN A. The non-Mendelian behavior of plant B chromosomes[J]. Chromosome Research, 2022, 30(2): 229-239. |
| [27] | KOO D H, HAN F P, BIRCHLER J A, et al. Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome[J]. Genome Research, 2011, 21(6): 908-914. |
| [28] | HUANG W, DU Y, ZHAO X, et al. B chromosome contains active genes and impacts the transcription of A chromosomes in maize (Zea mays L.)[J]. BMC Plant Biology, 2016, 16(1): 88. |
| [29] | BOUDICHEVSKAIA A, FIEBIG A, KUMKE K, et al. Rye B chromosomes differently influence the expression of A chromosome-encoded genes depending on the host species[J]. Chromosome Research, 2022, 30(4): 335-349. |
| [30] | LIU X, CHEN Z Q, SHI X W. The B chromosome: an optimum platform for maize minichromosome engineering[J]. Critical Reviews in Plant Sciences, 2025, 44(1): 30-45. |
| [31] | NELSON A D, LAMB J C, KOBROSSLY P S, et al. Parameters affecting telomere-mediated chromosomal truncation in Arabidopsis [J]. The Plant Cell, 2011, 23(6): 2263-2272. |
| [32] | TEO C H, MA L, KAPUSI E, et al. Induction of telomere-mediated chromosomal truncation and stability of truncated chromosomes in Arabidopsis thaliana [J]. The Plant Journal, 2011, 68(1): 28-39. |
| [33] | MURATA M. Artificial chromosome preparation in Arabidopsis [J]. Current Protocols in Plant Biology, 2016, 1(1): 53-66. |
| [34] | XU C H, CHENG Z K, YU W C. Construction of rice mini-chromosomes by telomere-mediated chromosomal truncation[J]. The Plant Journal, 2012, 70(6): 1070-1079. |
| [35] | YANG X Y, LI J H, CHEN L, et al. Stable mitotic inheritance of rice minichromosomes in cell suspension cultures[J]. Plant Cell Reports, 2015, 34(6): 929-941. |
| [36] | KAPUSI E, MA L, TEO C H, et al. Telomere-mediated truncation of barley chromosomes[J]. Chromosoma, 2012, 121(2): 181-190. |
| [37] | YUAN J, SHI Q H, GUO X, et al. Site-specific transfer of chromosomal segments and genes in wheat engineered chromosomes[J]. Journal of Genetics and Genomics, 2017, 44(11): 531-539. |
| [38] | YAN X H, LI C, YANG J, et al. Induction of telomere-mediated chromosomal truncation and behavior of truncated chromosomes in Brassica napus [J]. The Plant Journal, 2017, 91(4): 700-713. |
| [39] | YIN X Z, ZHANG Y X, CHEN Y H, et al. Precise characterization and tracking of stably inherited artificial minichromosomes made by telomere-mediated chromosome truncation in Brassica napus [J]. Frontiers in Plant Science, 2021, 12: 743792. |
| [40] | LIU Y, LIU Q, YI C Y, et al. Past innovations and future possibilities in plant chromosome engineering[J]. Plant Biotechnology Journal, 2025, 23(3): 695-708. |
| [41] | SEARS E R. The transfer of leaf-rust resistance from Aegilops umbellulata to wheat[J]. Genetics in plant breeding. Brook-haven Symposia in Biology, 1956, 9: 1-22. |
| [42] | FU S L, LV Z L, QI B, et al. Molecular cytogenetic characterization of wheat: Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium head blight[J]. Journal of Genetics and Genomics, 2012, 39(2): 103-110. |
| [43] | GUO X R, SHI Q H, LIU Y, et al. Systemic development of wheat-Thinopyrum elongatum translocation lines and their deployment in wheat breeding for Fusarium head blight resistance[J]. The Plant Journal, 2023, 114(6): 1475-1489. |
| [44] | KASZÁS E, BIRCHLER J A. Meiotic transmission rates correlate with physical features of rearranged centromeres in maize[J]. Genetics, 1998, 150(4): 1683-1692. |
| [45] | YAMADA T, FUJIMOTO Y, YAMAMOTO Y, et al. Minichromosome formation in Chlorella cells irradiated with electron beams[J]. Journal of Bioscience and Bioengineering, 2003, 95(6): 601-607. |
| [46] | LYZNIK L A, GORDON-KAMM W J, TAO Y. Site-specific recombination for genetic engineering in plants[J]. Plant Cell Reports, 2003, 21(10): 925-932. |
| [47] | SUN C, LEI Y, LI B S, et al. Precise integration of large DNA sequences in plant genomes using PrimeRoot editors[J]. Nature Biotechnology, 2024, 42(2): 316-327. |
| [48] | MURATA M, SHIBATA F, HIRONAKA A, et al. Generation of an artificial ring chromosome in Arabidopsis by Cre/LoxP-mediated recombination[J]. The Plant Journal, 2013, 74(3): 363-371. |
| [49] | MURATA M. Minichromosomes and artificial chromosomes in Arabidopsis [J]. Chromosome Research, 2014, 22(2): 167-178. |
| [50] | GAETA R T, MASONBRINK R E, ZHAO C Z, et al. In vivo modification of a maize engineered minichromosome[J]. Chromosoma, 2013, 122(3): 221-232. |
| [51] | BARRANGOU R, FREMAUX C, DEVEAU H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819): 1709-1712. |
| [52] | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| [53] | ZHOU H B, LIU B, WEEKS D P, et al. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice[J]. Nucleic Acids Research, 2014, 42(17): 10903-10914. |
| [54] | DURR J, PAPAREDDY R, NAKAJIMA K, et al. Highly efficient heritable targeted deletions of gene clusters and non-coding regulatory regions in Arabidopsis using CRISPR/Cas9[J]. Scientific Reports, 2018, 8: 4443. |
| [55] | LI R Q, CHAR S N, YANG B. Creating large chromosomal deletions in rice using CRISPR/Cas9[M/OL]//QI Y. Plant genome editing with CRISPR systems. New York: Springer New York, 2019: 47-61. (2019-01-05)[2025-09-01]. . |
| [56] | LI Y N, HUANG B Y, CHEN J, et al. Targeted large fragment deletion in plants using paired crRNAs with type Ⅰ CRISPR system[J]. Plant Biotechnology Journal, 2023, 21(11): 2196-2208. |
| [57] | RÖNSPIES M, SCHMIDT C, SCHINDELE P, et al. Massive crossover suppression by CRISPR-Cas-mediated plant chromosome engineering[J]. Nature Plants, 2022, 8(10): 1153-1159. |
| [58] | SCHMIDT C, PACHER M, PUCHTA H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system[J]. The Plant Journal, 2019, 98(4): 577-589. |
| [59] | SCHWARTZ C, LENDERTS B, FEIGENBUTZ L, et al. CRISPR-Cas9-mediated 75.5-Mb inversion in maize[J]. Nature Plants, 2020, 6(12): 1427-1431. |
| [60] | MAO Y H, ZHAO Y L, ZHOU Q, et al. Chromosome engineering: technologies, applications, and challenges[J]. Annual Review of Animal Biosciences, 2025, 13(1): 25-47. |
| [61] | WANG M L, LIN X J, MO B X, et al. Plant artificial chromosomes: construction and transformation[J]. ACS Synthetic Biology, 2024, 13(1): 15-24. |
| [62] | SHIZUYA H, KOUROS-MEHR H. The development and applications of the bacterial artificial chromosome cloning system[J]. The Keio Journal of Medicine, 2001, 50(1): 26-30. |
| [63] | HAO M L, TANG J B, GE S X, et al. Bacterial-artificial-chromosome-based genome editing methods and the applications in herpesvirus research[J]. Microorganisms, 2023, 11(3): 589. |
| [64] | KUDO K, NISHIMURA T, IZUMIKAWA M, et al. Capability of a large bacterial artificial chromosome clone harboring multiple biosynthetic gene clusters for the production of diverse compounds[J]. The Journal of Antibiotics, 2024, 77(5): 288-298. |
| [65] | MOGI K, TOMITA H, YOSHIHARA M, et al. Advances in bacterial artificial chromosome (BAC) transgenic mice for gene analysis and disease research[J]. Gene, 2025, 934: 149014. |
| [66] | CARLSON S R, RUDGERS G W, ZIELER H, et al. Meiotic transmission of an in vitro-assembled autonomous maize minichromosome[J]. PLoS Genetics, 2007, 3(10): e179. |
| [67] | ANANIEV E V, WU C C, CHAMBERLIN M A, et al. Artificial chromosome formation in maize (Zea mays L.)[J]. Chromosoma, 2009, 118(2): 157-177. |
| [68] | HAMILTON C M. A binary-BAC system for plant transformation with high-molecular-weight DNA[J]. Gene, 1997, 200(1-2): 107-116. |
| [69] | SHIBATA D, SEKI M, MITSUKAWA N, et al. Establishment of framework P1 clones for map-based cloning and genome sequencing: direct RFLP mapping of large clones[J]. Gene, 1998, 225(1-2): 31-38. |
| [70] | MOZO T, DEWAR K, DUNN P, et al. A complete BAC-based physical map of the Arabidopsis thaliana genome[J]. Nature Genetics, 1999, 22(3): 271-275. |
| [71] | LIU Y G, NAGAKI K, FUJITA M, et al. Development of an efficient maintenance and screening system for large-insert genomic DNA libraries of hexaploid wheat in a transformation-competent artificial chromosome (TAC) vector[J]. The Plant Journal, 2000, 23(5): 687-695. |
| [72] | LIU Y G, LIU H M, CHEN L T, et al. Development of new transformation-competent artificial chromosome vectors and rice genomic libraries for efficient gene cloning[J]. Gene, 2002, 282(1-2): 247-255. |
| [73] | HIROSE Y, SUDA K, LIU Y G, et al. The Arabidopsis TAC Position Viewer: a high-resolution map of transformation-competent artificial chromosome (TAC) clones aligned with the Arabidopsis thaliana Columbia-0 genome[J]. The Plant Journal, 2015, 83(6): 1114-1122. |
| [74] | QU S, COAKER G, FRANCIS D, et al. Development of a new transformation-competent artificial chromosome (TAC) vector and construction of tomato and rice TAC libraries[J]. Molecular Breeding, 2003, 12(4): 297-308. |
| [75] | LIANG F S, ZHANG K C, YU Z W, et al. Construction, characterization, and screening of a transformation-competent artificial chromosome library of peach[J]. Plant Molecular Biology Reporter, 2004, 22(1): 37-48. |
| [76] | KONG F N, JIANG S M, SHI L X, et al. Construction and characterization of a transformation-competent artificial chromosome (TAC) library of Zizania latifolia (Griseb.)[J]. Plant Molecular Biology Reporter, 2006, 24(2): 219-227. |
| [77] | WANG J Y, XIE Z X, CUI Y Z, et al. Artificial design of the genome: from sequences to the 3D structure of chromosomes[J]. Trends in Biotechnology, 2025, 43(2): 304-317. |
| [78] | CELLO J, PAUL A V, WIMMER E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template[J]. Science, 2002, 297(5583): 1016-1018. |
| [79] | SMITH H O, HUTCHISON C A, PFANNKOCH C, et al. Generating a synthetic genome by whole genome assembly: ϕX174 bacteriophage from synthetic oligonucleotides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(26): 15440-15445. |
| [80] | RICHARDSON S M, MITCHELL L A, STRACQUADANIO G, et al. Design of a synthetic yeast genome[J]. Science, 2017, 355(6329): 1040-1044. |
| [81] | THAO T THI NHU, LABROUSSAA F, EBERT N, et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform[J]. Nature, 2020, 582(7813): 561-565. |
| [82] | CHEN L G, LAN T L, ZHANG S, et al. A designer synthetic chromosome fragment functions in moss[J]. Nature Plants, 2024, 10(2): 228-239. |
| [83] | 朱骊宇, 赵玉龙, 李伟, 等. 哺乳动物染色体工程研究进展[J]. 合成生物学, 2023, 4(2): 394-406. |
| ZHU L Y, ZHAO Y L, LI W, et al. Progress in mammalian chromosome engineering[J]. Synthetic Biology Journal, 2023, 4(2): 394-406. | |
| [84] | ELENA C, RAVASI P, CASTELLI M E, et al. Expression of codon optimized genes in microbial systems: current industrial applications and perspectives[J]. Frontiers in Microbiology, 2014, 5: 21. |
| [85] | OSTROV N, LANDON M, GUELL M, et al. Design, synthesis, and testing toward a 57-codon genome[J]. Science, 2016, 353(6301): 819-822. |
| [86] | HOCHREIN L, MITCHELL L A, SCHULZ K, et al. L-SCRaMbLE as a tool for light-controlled Cre-mediated recombination in yeast[J]. Nature Communications, 2018, 9: 1931. |
| [87] | DYMOND J S, RICHARDSON S M, COOMBES C E, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design[J]. Nature, 2011, 477(7365): 471-476. |
| [88] | ANNALURU N, MULLER H, MITCHELL L A, et al. Total synthesis of a functional designer eukaryotic chromosome[J]. Science, 2014, 344(6179): 55-58. |
| [89] | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synⅡ, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355(6329): eaaf4791. |
| [90] | XIE Z X, LI B Z, MITCHELL L A, et al. “Perfect” designer chromosome Ⅴ and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| [91] | MITCHELL L A, WANG A, STRACQUADANIO G, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synⅥ and beyond[J]. Science, 2017, 355(6329): eaaf4831. |
| [92] | WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): eaaf4706. |
| [93] | SCHINDLER D, WALKER R S K, JIANG S Y, et al. Design, construction, and functional characterization of a tRNA neochromosome in yeast[J]. Cell, 2023, 186(24): 5237-5253.e22. |
| [94] | DU F, DAI J B, JIAO Y L. Insights into a functional synthetic plant genome[J]. New Phytologist, 2024, 244(1): 46-50. |
| [95] | YE H, LUO G Y, ZHENG Z W, et al. Plant synthetic genomics: big lessons from the little yeast[J]. Cell Chemical Biology, 2024, 31(10): 1745-1754. |
| [96] | YU W F, ZHANG S, ZHAO S J, et al. Designing a synthetic moss genome using GenoDesigner[J]. Nature Plants, 2024, 10(6): 848-856. |
| [97] | LANG D, ULLRICH K K, MURAT F, et al. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution[J]. The Plant Journal, 2018, 93(3): 515-533. |
| [98] | BI G Q, ZHAO S J, YAO J W, et al. Near telomere-to-telomere genome of the model plant Physcomitrium patens [J]. Nature Plants, 2024, 10(2): 327-343. |
| [99] | ISAACS F J, CARR P A, WANG H H, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement[J]. Science, 2011, 333(6040): 348-353. |
| [100] | FREDENS J, WANG K H, DE LA TORRE D, et al. Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569(7757): 514-518. |
| [101] | ROVNER A J, HAIMOVICH A D, KATZ S R, et al. Recoded organisms engineered to depend on synthetic amino acids[J]. Nature, 2015, 518(7537): 89-93. |
| [102] | CHENG L, ZHAO S J, LI T Y, et al. Large-scale genomic rearrangements boost SCRaMbLE in Saccharomyces cerevisiae [J]. Nature Communications, 2024, 15: 770. |
| [103] | BLOUNT B A, GOWERS G F, HO J C H, et al. Rapid host strain improvement by in vivo rearrangement of a synthetic yeast chromosome[J]. Nature Communications, 2018, 9: 1932. |
| [104] | LUO Z Q, WANG L H, WANG Y, et al. Identifying and characterizing SCRaMbLEd synthetic yeast using ReSCuES[J]. Nature Communications, 2018, 9: 1930. |
| [105] | JIA B, WU Y, LI B Z, et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nature Communications, 2018, 9: 1933. |
| [106] | SCHINDLER D, DAI J B, CAI Y Z. Synthetic genomics: a new venture to dissect genome fundamentals and engineer new functions[J]. Current Opinion in Chemical Biology, 2018, 46: 56-62. |
| [107] | JIANG S Y, TANG Y W, XIANG L, et al. Efficient de novo assembly and modification of large DNA fragments[J]. Science China Life Sciences, 2022, 65(7): 1445-1455. |
| [108] | SHAO Y Y, LU N, WU Z F, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| [109] | GU X, YE T T, ZHANG X R, et al. Single-chromosome fission yeast models reveal the configuration robustness of a functional genome[J]. Cell Reports, 2022, 40(8): 111237. |
| [110] | WANG L B, LI Z K, WANG L Y, et al. A sustainable mouse karyotype created by programmed chromosome fusion[J]. Science, 2022, 377(6609): 967-975. |
| [111] | MERCY G, MOZZICONACCI J, SCOLARI V F, et al. 3D organization of synthetic and scrambled chromosomes[J]. Science, 2017, 355(6329): eaaf4597. |
| [112] | ZHANG W M, LAZAR-STEFANITA L, YAMASHITA H, et al. Manipulating the 3D organization of the largest synthetic yeast chromosome[J]. Molecular Cell, 2023, 83(23): 4424-4437.e5. |
| [113] | WESTHORPE F G, STRAIGHT A F. Chromosome segregation: reconstituting the kinetochore[J]. Current Biology, 2016, 26(23): R1242-R1245. |
| [114] | EVATT J M, SADLI A D, RAPACZ B K, et al. Centromere pairing enables correct segregation of meiotic chromosomes[J]. Current Biology, 2024, 34(10): 2085-2093.e6. |
| [115] | CLEVELAND D W, MAO Y H, SULLIVAN K F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling[J]. Cell, 2003, 112(4): 407-421. |
| [116] | PROSÉE R F, WENDA J M, STEINER F A. Adaptations for centromere function in meiosis[J]. Essays in Biochemistry, 2020, 64(2): 193-203. |
| [117] | NAISH M, HENDERSON I R. The structure, function, and evolution of plant centromeres[J]. Genome Research, 2024, 34(2): 161-178. |
| [118] | KOCH L B, MARSTON A L. The functional organisation of the centromere and kinetochore during meiosis[J]. Current Opinion in Cell Biology, 2025, 94: 102486. |
| [119] | MELTERS D P, BRADNAM K R, YOUNG H A, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution[J]. Genome Biology, 2013, 14(1): R10. |
| [120] | WLODZIMIERZ P, RABANAL F A, BURNS R, et al. Cycles of satellite and transposon evolution in Arabidopsis centromeres[J]. Nature, 2023, 618(7965): 557-565. |
| [121] | LOGSDON G A, ROZANSKI A N, RYABOV F, et al. The variation and evolution of complete human centromeres[J]. Nature, 2024, 629(8010): 136-145. |
| [122] | COPENHAVER G P, NICKEL K, KUROMORI T, et al. Genetic definition and sequence analysis of Arabidopsis centromeres[J]. Science, 1999, 286(5449): 2468-2474. |
| [123] | NAGAKI K, CHENG Z K, OUYANG S, et al. Sequencing of a rice centromere uncovers active genes[J]. Nature Genetics, 2004, 36(2): 138-145. |
| [124] | NAISH M, ALONGE M, WLODZIMIERZ P, et al. The genetic and epigenetic landscape of the Arabidopsis centromeres[J]. Science, 2021, 374(6569): eabi7489. |
| [125] | CHENG Z K, DONG F G, LANGDON T, et al. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon[J]. The Plant Cell, 2002, 14(8): 1691-1704. |
| [126] | ZHONG C X, MARSHALL J B, TOPP C, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3[J]. The Plant Cell, 2002, 14(11): 2825-2836. |
| [127] | NAGAKI K, NEUMANN P, ZHANG D F, et al. Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice[J]. Molecular Biology and Evolution, 2005, 22(4): 845-855. |
| [128] | SONG J M, XIE W Z, WANG S, et al. Two gap-free reference genomes and a global view of the centromere architecture in rice[J]. Molecular Plant, 2021, 14(10): 1757-1767. |
| [129] | CHEN J, WANG Z J, TAN K W, et al. A complete telomere-to-telomere assembly of the maize genome[J]. Nature Genetics, 2023, 55(7): 1221-1231. |
| [130] | CLARKE L, CARBON J. Isolation of a yeast centromere and construction of functional small circular chromosomes[J]. Nature, 1980, 287(5782): 504-509. |
| [131] | KOUPRINA N, EBERSOLE T, KORIABINE M, et al. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes[J]. Nucleic Acids Research, 2003, 31(3): 922-934. |
| [132] | TALBERT P B, MASUELLI R, TYAGI A P, et al. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant[J]. The Plant Cell, 2002, 14(5): 1053-1066. |
| [133] | HAN F P, LAMB J C, BIRCHLER J A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(9): 3238-3243. |
| [134] | PHAN B H, JIN W W, TOPP C N, et al. Transformation of rice with long DNA-segments consisting of random genomic DNA or centromere-specific DNA[J]. Transgenic Research, 2007, 16(3): 341-351. |
| [135] | HARA M, FUKAGAWA T. Where is the right path heading from the centromere to spindle microtubules?[J]. Cell Cycle, 2019, 18(11): 1199-1211. |
| [136] | RAIPURIA R K, WATTS A, SHARMA B B, et al. Decoding allelic diversity, transcript variants and transcriptional complexity of CENH3 gene in Brassica oleracea var. botrytis [J]. Protoplasma, 2023, 260(4): 1149-1162. |
| [137] | LERMONTOVA I, SCHUBERT V, FUCHS J, et al. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain[J]. The Plant Cell, 2006, 18(10): 2443-2451. |
| [138] | LERMONTOVA I, KOROLEVA O, RUTTEN T, et al. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation[J]. The Plant Journal, 2011, 68(1): 40-50. |
| [139] | KARIMI-ASHTIYANI R, ISHII T, NIESSEN M, et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(36): 11211-11216. |
| [140] | RAVI M, CHAN S W L. Haploid plants produced by centromere-mediated genome elimination[J]. Nature, 2010, 464(7288): 615-618. |
| [141] | KELLIHER T, STARR D, WANG W L, et al. Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize[J]. Frontiers in Plant Science, 2016, 7: 414. |
| [142] | RAVI M, SHIBATA F, RAMAHI J S, et al. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana [J]. PLoS Genetics, 2011, 7(6): e1002121. |
| [143] | WANG N, DAWE R K. Centromere size and its relationship to haploid formation in plants[J]. Molecular Plant, 2018, 11(3): 398-406. |
| [144] | DUNLEAVY E M, ROCHE D, TAGAMI H, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres[J]. Cell, 2009, 137(3): 485-497. |
| [145] | FOLTZ D R, JANSEN L E T, BAILEY A O, et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP[J]. Cell, 2009, 137(3): 472-484. |
| [146] | MENDIBURO M J, PADEKEN J, FÜLÖP S, et al. Drosophila CENH3 is sufficient for centromere formation[J]. Science, 2011, 334(6056): 686-690. |
| [147] | LOGSDON G A, GAMBOGI C W, LISKOVYKH M A, et al. Human artificial chromosomes that bypass centromeric DNA[J]. Cell, 2019, 178(3): 624-639.e19. |
| [148] | GAMBOGI C W, BIRCHAK G J, MER E, et al. Efficient formation of single-copy human artificial chromosomes[J]. Science, 2024, 383(6689): 1344-1349. |
| [149] | TEO C H, LERMONTOVA I, HOUBEN A, et al. De novo generation of plant centromeres at tandem repeats[J]. Chromosoma, 2013, 122(3): 233-241. |
| [150] | DAWE R K, GENT J I, ZENG Y B, et al. Synthetic maize centromeres transmit chromosomes across generations[J]. Nature Plants, 2023, 9(3): 433-441. |
| [151] | ZENG Y B, WANG M Y, GENT J I, et al. Increased maize chromosome number by engineered chromosome fission[J]. Science Advances, 2025, 11(21): eadw3433. |
| [152] | BAI S, LUO H, TONG H Z, et al. Advances on transfer and maintenance of large DNA in bacteria, fungi, and mammalian cells[J]. Biotechnology Advances, 2024, 76: 108421. |
| [153] | CAMPELO S N, HUANG P H, BUIE C R, et al. Recent advancements in electroporation technologies: from bench to clinic[J]. Annual Review of Biomedical Engineering, 2023, 25: 77-100. |
| [154] | LEE E C, LIANG Q, ALI H, et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery[J]. Nature Biotechnology, 2014, 32(4): 356-363. |
| [155] | MITCHELL L A, MCCULLOCH L H, PINGLAY S, et al. De novo assembly and delivery to mouse cells of a 101 kb functional human gene[J]. Genetics, 2021, 218(1): iyab038. |
| [156] | MARSCHALL P, MALIK N, LARIN Z. Transfer of YACs up to 2.3 Mb intact into human cells with polyethylenimine[J]. Gene Therapy, 1999, 6(9): 1634-1637. |
| [157] | MEJÍA J E, WILLMOTT A, LEVY E, et al. Functional complementation of a genetic deficiency with human artificial chromosomes[J]. The American Journal of Human Genetics, 2001, 69(2): 315-326. |
| [158] | MARTINEZ A, KOLVEK S J, YIP C L T, et al. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts[J]. Applied and Environmental Microbiology, 2004, 70(4): 2452-2463. |
| [159] | BAÑUELOS-VAZQUEZ L A, TORRES TEJERIZO G, BROM S. Regulation of conjugative transfer of plasmids and integrative conjugative elements[J]. Plasmid, 2017, 91: 82-89. |
| [160] | BROPHY J A N, TRIASSI A J, ADAMS B L, et al. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria[J]. Nature Microbiology, 2018, 3(9): 1043-1053. |
| [161] | LI L P, BLANKENSTEIN T. Generation of transgenic mice with megabase-sized human yeast artificial chromosomes by yeast spheroplast-embryonic stem cell fusion[J]. Nature Protocols, 2013, 8(8): 1567-1582. |
| [162] | BROWN D M, CHAN Y A, DESAI P J, et al. Efficient size-independent chromosome delivery from yeast to cultured cell lines[J]. Nucleic Acids Research, 2017, 45(7): e50. |
| [163] | EGE T, RINGERTZ N R. Preparation of microcells by enucleation of micronucleate cells[J]. Experimental Cell Research, 1974, 87(2): 378-382. |
| [164] | O’DOHERTY A, RUF S, MULLIGAN C, et al. An aneuploid mouse strain carrying human chromosome 21 with down syndrome phenotypes[J]. Science, 2005, 309(5743): 2033-2037. |
| [165] | KATOH M, KAZUKI Y, KAZUKI K, et al. Exploitation of the interaction of measles virus fusogenic envelope proteins with the surface receptor CD46 on human cells for microcell-mediated chromosome transfer[J]. BMC Biotechnology, 2010, 10: 37. |
| [166] | LISKOVYKH M, LEE N C O, LARIONOV V, et al. Moving toward a higher efficiency of microcell-mediated chromosome transfer[J]. Molecular Therapy - Methods & Clinical Development, 2016, 3: 16043. |
| [167] | MIYAMOTO H, KOBAYASHI H, KISHIMA N, et al. Rapid human genomic DNA cloning into mouse artificial chromosome via direct chromosome transfer from human iPSC and CRISPR/Cas9-mediated translocation[J]. Nucleic Acids Research, 2024, 52(3): 1498-1511. |
| [168] | WANG Y F, ZENG H Y, ZHOU X, et al. Transformation of rice with large maize genomic DNA fragments containing high content repetitive sequences[J]. Plant Cell Reports, 2015, 34(6): 1049-1061. |
| [169] | MULLEN J, ADAM G, BLOWERS A, et al. Biolistic transfer of large DNA fragments to tobacco cells using YACs retrofitted for plant transformation[J]. Molecular Breeding, 1998, 4(5): 449-457. |
| [170] | ZHAO X, MENG Z G, WANG Y, et al. Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers[J]. Nature Plants, 2017, 3(12): 956-964. |
| [171] | VAN WORDRAGEN M, SHAKYA R, VERKERK R, et al. Liposome-mediated transfer of YAC-DNA to tobacco cells[J]. Plant Molecular Biology Reporter, 1997, 15(2): 170-178. |
| [172] | 逄春梅, 张亮, 孙春玉, 等. 人参大片段DNA(100kb)转化灵芝的研究[J]. 菌物学报, 2013, 32(1): 96-102. |
| PANG C M, ZHANG L, SUN C Y, et al. Transformation of large DNA fragment of Panax ginseng into Ganoderma lucidum [J]. Mycosystema, 2013, 32(1): 96-102. | |
| [173] | HAMILTON C M, FRARY A, XU Y M, et al. Construction of tomato genomic DNA libraries in a binary-BAC (BIBAC) vector[J]. The Plant Journal, 1999, 18(2): 223-229. |
| [174] | SHIBATA D, LIU Y G. Technical focus-Agrobacterium-mediated plant transformation with large DNA fragments[J]. Trends in Plant Science, 2000, 5(8): 354-357. |
| [175] | VEGA J M, YU W C, HAN F P, et al. Agrobacterium-mediated transformation of maize (Zea mays) with Cre-lox site specific recombination cassettes in BIBAC vectors[J]. Plant Molecular Biology, 2008, 66(6): 587-598. |
| [176] | HE R F, WANG Y Y, SHI Z Y, et al. Construction of a genomic library of wild rice and Agrobacterium-mediated transformation of large insert DNA linked to BPH resistance locus[J]. Gene, 2003, 321: 113-121. |
| [177] | HE R F, PAN J, ZHU L L, et al. Agrobacterium-mediated transformation of large DNA fragments using a BIBAC vector system in rice[J]. Plant Molecular Biology Reporter, 2010, 28(4): 613-619. |
| [178] | KLEIN T M, WOLF E D, WU R, et al. High-velocity microprojectiles for delivering nucleic acids into living cells[J]. Nature, 1987, 327(6117): 70-73. |
| [179] | ALTPETER F, BAISAKH N, BEACHY R, et al. Particle bombardment and the genetic enhancement of crops: myths and realities[J]. Molecular Breeding, 2005, 15(3): 305-327. |
| [180] | LACROIX B, CITOVSKY V. Biolistic approach for transient gene expression studies in plants[M/OL]//RUSTGI S, LUO H. Methods in molecular biology: biolistic DNA delivery in plants. New York: Springer US, 2020: 125-139. (2020-04-11)[2025-09-01]. . |
| [181] | ERCOLANO M R, BALLVORA A, PAAL J, et al. Functional complementation analysis in potato via biolistic transformation with BAC large DNA fragments[J]. Molecular Breeding, 2004, 13(1): 15-22. |
| [182] | CHANG Y L, CHUANG H W, MEKSEM K, et al. Characterization of a plant-transformation-ready large-insert BIBAC library of Arabidopsis and bombardment transformation of a large-insert BIBAC of the library into tobacco[J]. Genome, 2011, 54(6): 437-447. |
| [183] | CUNNINGHAM F J, DEMIRER G S, GOH N S, et al. Nanobiolistics: an emerging genetic transformation approach[M/OL]//RUSTGI S, LUO H. Methods in molecular biology: biolistic DNA delivery in plants. New York: Springer US, 2020: 141-159. (2020-04-11)[2025-09-01]. . |
| [184] | LV Z Y, JIANG R, CHEN J F, et al. Nanoparticle-mediated gene transformation strategies for plant genetic engineering[J]. The Plant Journal, 2020, 104(4): 880-891. |
| [185] | DEMIRER G S, ZHANG H, GOH N S, et al. Carbon nanotube-mediated DNA delivery without transgene integration in intact plants[J]. Nature Protocols, 2019, 14(10): 2954-2971. |
| [186] | MORGAN J M, JELENSKA J, HENSLEY D, et al. An efficient and broadly applicable method for transient transformation of plants using vertically aligned carbon nanofiber arrays[J]. Frontiers in Plant Science, 2022, 13: 1051340. |
| [187] | TORNEY F, TREWYN B G, LIN V S Y, et al. Mesoporous silica nanoparticles deliver DNA and chemicals into plants[J]. Nature Nanotechnology, 2007, 2(5): 295-300. |
| [188] | CUNNINGHAM F J, GOH N S, DEMIRER G S, et al. Nanoparticle-mediated delivery towards advancing plant genetic engineering[J]. Trends in Biotechnology, 2018, 36(9): 882-897. |
| [189] | TERAMURA Y, KANEDA Y, TOTANI T, et al. Behavior of synthetic polymers immobilized on a cell membrane[J]. Biomaterials, 2008, 29(10): 1345-1355. |
| [190] | ZHENG H Z, LIU H H, CHEN S X, et al. Yeast transformation process studied by fluorescence labeling technique[J]. Bioconjugate Chemistry, 2005, 16(2): 250-254. |
| [191] | HASEZAWA S, MATSUI C, NAGATA T, et al. Cytological study of the introduction of Agrobacterium tumefaciens spheroplasts into Vinca rosea protoplasts[J]. Canadian Journal of Botany, 1983, 61(4): 1052-1057. |
| [192] | HATSUYAMA Y, SUNAGA N, HABU Y, et al. Direct transfer of plasmid DNA from intact yeast spheroplasts into plant protoplasts[J]. Plant & Cell Physiology, 1994, 35(1): 93-98. |
| [193] | YADAV T, QUIVY J P, ALMOUZNI G. Chromatin plasticity: a versatile landscape that underlies cell fate and identity[J]. Science, 2018, 361(6409): 1332-1336. |
| [194] | BARNHART M C, KUICH P H J L, STELLFOX M E, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore[J]. The Journal of Cell Biology, 2011, 194(2): 229-243. |
| [195] | CAMAHORT R, LI B, FLORENS L, et al. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore[J]. Molecular Cell, 2007, 26(6): 853-865. |
| [196] | DUNLEAVY E M, PIDOUX A L, MONET M, et al. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres[J]. Molecular Cell, 2007, 28(6): 1029-1044. |
| [197] | STOLER S, ROGERS K, WEITZE S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(25): 10571-10576. |
| [198] | WILLIAMS J S, HAYASHI T, YANAGIDA M, et al. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin[J]. Molecular Cell, 2009, 33(3): 287-298. |
| [199] | CHEN C C, DECHASSA M L, BETTINI E, et al. CAL1 is the Drosophila CENP-A assembly factor[J]. The Journal of Cell Biology, 2014, 204(3): 313-329. |
| [200] | LERMONTOVA I, KUHLMANN M, FRIEDEL S, et al. Arabidopsis KINETOCHORE NULL2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres[J]. The Plant Cell, 2013, 25(9): 3389-3404. |
| [201] | ZUO S, YADALA R, YANG F, et al. Recurrent plant-specific duplications of KNL2 and its conserved function as a kinetochore assembly factor[J]. Molecular Biology and Evolution, 2022, 39(6): msac123. |
| [202] | WOO H R, PONTES O, PIKAARD C S, et al. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization[J]. Genes & Development, 2007, 21(3): 267-277. |
| [203] | KRAFT E, BOSTICK M, JACOBSEN S E, et al. ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases[J]. The Plant Journal, 2008, 56(5): 704-715. |
| [204] | ZÜRCHER J F, KLEEFELDT A A, FUNKE L F H, et al. Continuous synthesis of E. coli genome sections and Mb-scale human DNA assembly[J]. Nature, 2023, 619(7970): 555-562. |
| [205] | ZHONG L, ZHANG Q, LU N, et al. The conjugation-associated linear-BAC iterative assembling (CALBIA) method for cloning 2.1-Mb human chromosomal DNAs in bacteria[J]. Cell Research, 2025, 35(4): 309-312. |
| [206] | LAN T L, CHEN L G, WANG Y, et al. Genome synthesis in plants[J]. Nature Reviews Bioengineering, 2025: 326. |
| [207] | SHAKIROV E V, CHEN J J L, SHIPPEN D E. Plant telomere biology: the green solution to the end-replication problem[J]. The Plant Cell, 2022, 34(7): 2492-2504. |
| [208] | MCKNIGHT T D, RIHA K, SHIPPEN D E. Telomeres, telomerase, and stability of the plant genome[J]. Plant Molecular Biology, 2002, 48(4): 331-337. |
| [209] | HOWELL S H. Molecular biology[M/OL]. New York: Springer New York, 2014[2025-09-01]. . |
| [210] | HU Y X, STILLMAN B. Origins of DNA replication in eukaryotes[J]. Molecular Cell, 2023, 83(3): 352-372. |
| [211] | STILLMAN B, DIFFLEY J F X, IWASA J H. Mechanisms for licensing origins of DNA replication in eukaryotic cells[J]. Nature Structural & Molecular Biology, 2025, 32(7): 1143-1153. |
| [212] | WHEELER E, BROOKS A M, CONCIA L, et al. Arabidopsis DNA replication initiates in intergenic, AT-rich open chromatin[J]. Plant Physiology, 2020, 183(1): 206-220. |
| [213] | FIORUCCI A S. AT the onset of DNA replication in Arabidopsis [J]. Plant Physiology, 2020, 183(1): 19-20. |
| [214] | JACOB Y, STROUD H, LEBLANC C, et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases[J]. Nature, 2010, 466(7309): 987-991. |
| [215] | COSTAS C, DE LA PAZ SANCHEZ M, STROUD H, et al. Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks[J]. Nature Structural & Molecular Biology, 2011, 18(3): 395-400. |
| [216] | STROUD H, HALE C J, FENG S H, et al. DNA methyltransferases are required to induce heterochromatic re-replication in Arabidopsis [J]. PLoS Genetics, 2012, 8(7): e1002808. |
| [217] | EBERSOLE T A, ROSS A, CLARK E, et al. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats[J]. Human Molecular Genetics, 2000, 9(11): 1623-1631. |
| [218] | OKADA T, OHZEKI J I, NAKANO M, et al. CENP-B controls centromere formation depending on the chromatin context[J]. Cell, 2007, 131(7): 1287-1300. |
| [219] | NAKANO M, CARDINALE S, NOSKOV V N, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers[J]. Developmental Cell, 2008, 14(4): 507-522. |
| [220] | WEUTS A, VOET T, VERBEECK J, et al. Telomere length homeostasis and telomere position effect on a linear human artificial chromosome are dictated by the genetic background[J]. Nucleic Acids Research, 2012, 40(22): 11477-11489. |
| [221] | KOUPRINA N, PETROV N, MOLINA O, et al. Human artificial chromosome with regulated centromere: a tool for genome and cancer studies[J]. ACS Synthetic Biology, 2018, 7(9): 1974-1989. |
| [222] | ZHU X T, LIU S C, YE T T, et al. Artificial chromosome reorganization reveals high plasticity of the budding and fission yeast genomes[J]. Genome Biology, 2025, 26(1): 229. |
| [223] | D’HALLUIN K, VANDERSTRAETEN C, VAN HULLE J, et al. Targeted molecular trait stacking in cotton through targeted double-strand break induction[J]. Plant Biotechnology Journal, 2013, 11(8): 933-941. |
| [224] | HOU L L, YAU Y Y, WEI J J, et al. An open-source system for in planta gene stacking by Bxb1 and Cre recombinases[J]. Molecular Plant, 2014, 7(12): 1756-1765. |
| [225] | KUMAR S, ALABED D, WORDEN A, et al. A modular gene targeting system for sequential transgene stacking in plants[J]. Journal of Biotechnology, 2015, 207: 12-20. |
| [226] | SRIVASTAVA V, THOMSON J. Gene stacking by recombinases[J]. Plant Biotechnology Journal, 2016, 14(2): 471-482. |
| [227] | ANAND A, WU E, LI Z, et al. High efficiency Agrobacterium-mediated site-specific gene integration in maize utilizing the FLP-FRT recombination system[J]. Plant Biotechnology Journal, 2019, 17(8): 1636-1645. |
| [228] | LI Y M, LI R Y, HAN Z G, et al. Recombinase-mediated gene stacking in cotton[J]. Plant Physiology, 2022, 188(4): 1852-1865. |
| [229] | ZONG Y, LIU Y J, XUE C X, et al. An engineered prime editor with enhanced editing efficiency in plants[J]. Nature Biotechnology, 2022, 40(9): 1394-1402. |
| [230] | LIU P, PANDA K, EDWARDS S A, et al. Transposase-assisted target-site integration for efficient plant genome engineering[J]. Nature, 2024, 631(8021): 593-600. |
| [231] | SUN C, LI H C, LIU Y J, et al. Iterative recombinase technologies for efficient and precise genome engineering across kilobase to megabase scales[J]. Cell, 2025, 188(17): 4693-4710.e15. |
| [232] | ELSHAMEY E A, YANG X M, YANG J Z, et al. Occurrence, biosynthesis, and health benefits of anthocyanins in rice and barley[J]. International Journal of Molecular Sciences, 2025, 26(13): 6225. |
| [233] | YE X D, AL-BABILI S, KLÖTI A, et al. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm[J]. Science, 2000, 287(5451): 303-305. |
| [234] | PAINE J A, SHIPTON C A, CHAGGAR S, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content[J]. Nature Biotechnology, 2005, 23(4): 482-487. |
| [235] | LUDWIG M. Evolution of the C4 photosynthetic pathway: events at the cellular and molecular levels[J]. Photosynthesis Research, 2013, 117(1): 147-161. |
| [236] | QIN K Z, YE X Y, LUO S S, et al. Engineering carbon assimilation in plants[J]. Journal of Integrative Plant Biology, 2025, 67(4): 926-948. |
| [237] | GUO K Y, YANG J, YU N, et al. Biological nitrogen fixation in cereal crops: Progress, strategies, and perspectives[J]. Plant Communications, 2023, 4(2): 100499. |
| [238] | LIU F, ZHAO Z H, FERNIE A R, et al. Towards establishing functional nitrogenase activities within plants[J/OL]. Trends in Biotechnology, 2025. (2025-05-28)[2025-09-01]. . |
| [239] | WANG J, XIE Z X, MA Y, et al. Ring synthetic chromosome Ⅴ SCRaMbLE[J]. Nature Communications, 2018, 9: 3783. |
| [240] | CHOI S, NAH H J, CHOI S, et al. Heterologous expression of daptomycin biosynthetic gene cluster via Streptomyces artificial chromosome vector system[J]. Journal of Microbiology and Biotechnology, 2019, 29(12): 1931-1937. |
| [241] | TIAN T, WU X W, WU P P, et al. High-level expression of leghemoglobin in Kluyveromyces marxianus by remodeling the heme metabolism pathway[J]. Frontiers in Bioengineering and Biotechnology, 2024, 11: 1329016. |
| [242] | TRIPATHY S, DASSARMA B, BHATTACHARYA M, et al. Plant-based vaccine research development against viral diseases with emphasis on Ebola virus disease: a review study[J]. Current Opinion in Pharmacology, 2021, 60: 261-267. |
| [243] | JUGLER C, SUN H Y, NGUYEN K, et al. A novel plant-made monoclonal antibody enhances the synergetic potency of an antibody cocktail against the SARS-CoV-2 Omicron variant[J]. Plant Biotechnology Journal, 2023, 21(3): 549-559. |
| [1] | WEI Jiaxiu, JI Peiyun, JIE Qingyu, HUANG Qiuyan, YE Hao, DAI Junbiao. Construction and application of plant artificial chromosomes [J]. Synthetic Biology Journal, 2025, 6(5): 1093-1106. |
| [2] | YUAN Shengjian, MA Yingfei. Advances and applications of phage synthetic biology [J]. Synthetic Biology Journal, 2020, 1(6): 635-655. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||