Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (5): 1093-1106.DOI: 10.12211/2096-8280.2025-086
• Invited Review • Previous Articles Next Articles
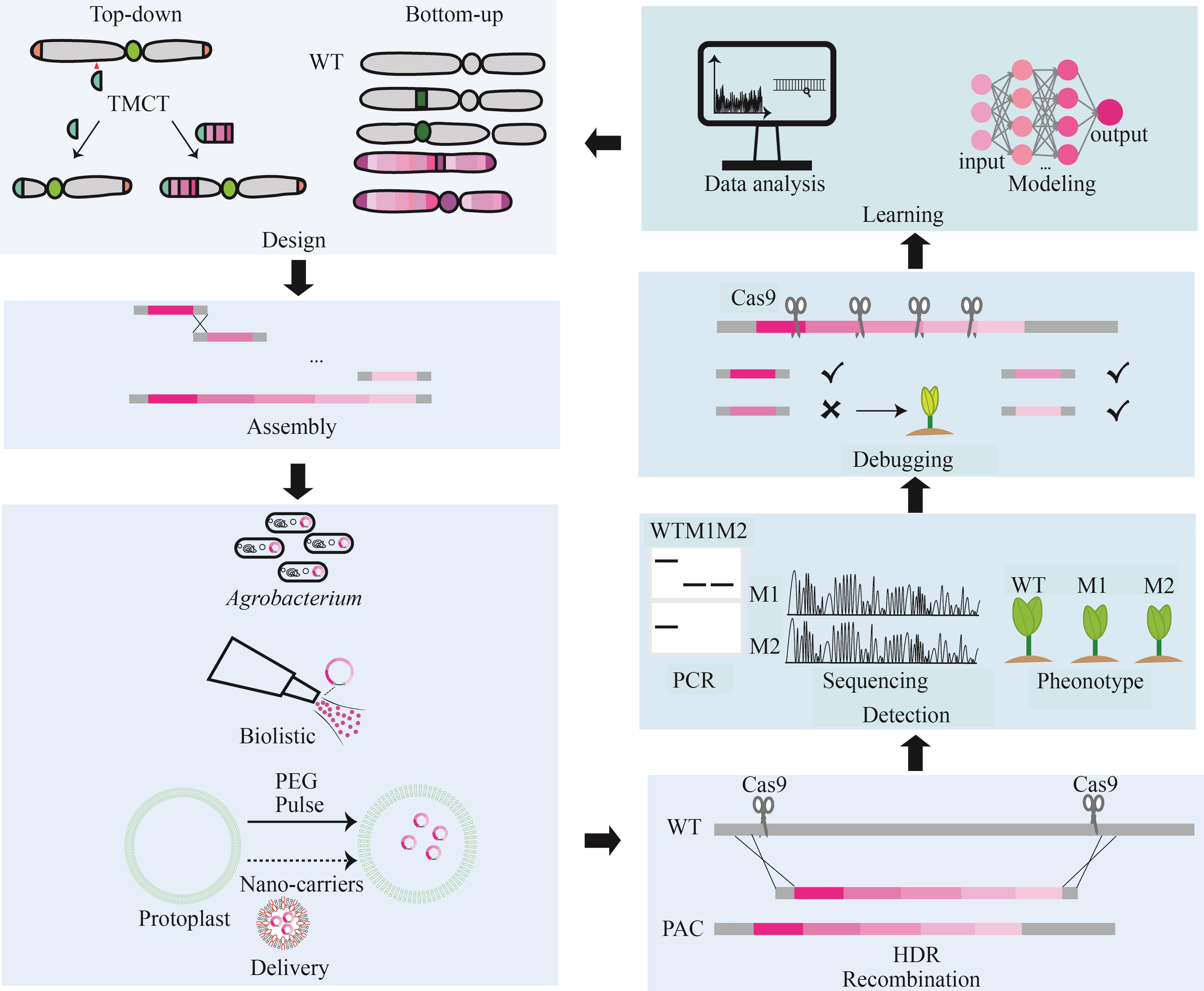
Construction and application of plant artificial chromosomes
WEI Jiaxiu1, JI Peiyun1, JIE Qingyu1, HUANG Qiuyan1, YE Hao1, DAI Junbiao1,2
- 1.Shenzhen Branch,Guangdong Laboratory of Lingnan Modern Agriculture,Key Laboratory of Synthetic Biology,Ministry of Agriculture and Rural Affairs,Agricultural Genomics Institute at Shenzhen,Chinese Academy of Agricultural Sciences,Shenzhen 518120,Guangdong,China
2.Guangdong Provincial Key Laboratory of Synthetic Genomics,Shenzhen Key Laboratory of Synthetic Genomics,Key Laboratory of Quantitative Synthetic Biology,Shenzhen Institute of Synthetic Biology,Shenzhen Institute of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
-
Received:2025-08-20Revised:2025-09-27Online:2025-11-05Published:2025-10-31 -
Contact:DAI Junbiao
植物人工染色体的构建与应用
魏家秀1, 嵇佩云1, 节庆雨1, 黄秋燕1, 叶浩1, 戴俊彪1,2
- 1.中国农业科学院农业基因组研究所,农业农村部合成生物学重点实验室,广东省岭南现代农业实验室,广东 深圳 518120
2.中国科学院深圳先进技术研究院合成生物学研究所,广东省合成基因组学重点实验室,深圳市合成基因组学重点实验室,定量合成生物学重点实验室,广东 深圳 518055
-
通讯作者:戴俊彪 -
作者简介:魏家秀 (1992—),女,博士。研究方向为叶绿体基因组的核迁移。E-mail:weijiaxiu@caas.cn嵇佩云 (1996—),女,博士。研究方向为植物高效同源重组元件的挖掘与应用。E-mail:jipeiyun@caas.cn戴俊彪 (1974—),男,研究员,博士生导师,广东省合成基因组学重点实验室主任,深圳合成基因组学重点实验室主任,欧洲科学院院士。研究方向为开发基因和基因组的合成、组装及转移技术,通过基因组的设计构建解析基因组功能,并进行合成生物的改造和优化等方面的研究工作。E-mail:daijunbiao@caas.cn -
基金资助:国家重点研发计划(2023YFA0913500);国家自然科学基金(32301232);中国农业科学院科技创新工程专项(2060299)
CLC Number:
Cite this article
WEI Jiaxiu, JI Peiyun, JIE Qingyu, HUANG Qiuyan, YE Hao, DAI Junbiao. Construction and application of plant artificial chromosomes[J]. Synthetic Biology Journal, 2025, 6(5): 1093-1106.
魏家秀, 嵇佩云, 节庆雨, 黄秋燕, 叶浩, 戴俊彪. 植物人工染色体的构建与应用[J]. 合成生物学, 2025, 6(5): 1093-1106.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-086
| [1] | COHEN S N, CHANG A C Y, BOYER H W, et al. Construction of biologically functional bacterial plasmids in vitro [J]. Proceedings of the National Academy of Sciences of the United States of America, 1973, 70(11): 3240-3244. |
| [2] | MURRAY A W, SZOSTAK J W. Construction of artificial chromosomes in yeast[J]. Nature, 1983, 305(5931): 189-193. |
| [3] | BURKE D T, CARLE G F, OLSON M V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors[J]. Science, 1987, 236(4803): 806-812. |
| [4] | HARRINGTON J J, BOKKELEN G V, MAYS R W, et al. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes[J]. Nature Genetics, 1997, 15(4): 345-355. |
| [5] | CARLSON S R, RUDGERS G W, ZIELER H, et al. Meiotic transmission of an in vitro-assembled autonomous maize minichromosome[J]. PLoS Genetics, 2007, 3(10): e179. |
| [6] | YU W C, LAMB J C, HAN F P, et al. Telomere-mediated chromosomal truncation in maize[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(46): 17331-17336. |
| [7] | BIRCHLER J A, HAN F P. Maize centromeres: structure, function, epigenetics[J]. Annual Review of Genetics, 2009, 43: 287-303. |
| [8] | JIANG S Y, LUO Z Q, WU J, et al. Building a eukaryotic chromosome arm by de novo design and synthesis[J]. Nature Communications, 2023, 14: 7886. |
| [9] | JAKUBIEC A, SAROKINA A, CHOINARD S, et al. Replicating minichromosomes as a new tool for plastid genome engineering[J]. Nature Plants, 2021, 7(7): 932-941. |
| [10] | SHAO R F, BARKER S C. Chimeric mitochondrial minichromosomes of the human body louse, Pediculus humanus: evidence for homologous and non-homologous recombination[J]. Gene, 2011, 473(1): 36-43. |
| [11] | RUF S, KARCHER D, BOCK R. Determining the transgene containment level provided by chloroplast transformation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(17): 6998-7002. |
| [12] | MALIGA P. Plastid transformation in higher plants[J]. Annual Review of Plant Biology, 2004, 55: 289-313. |
| [75] | LIU Y G, SHIRANO Y, FUKAKI H, et al. Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(11): 6535-6540. |
| [76] | LIN L, LIU Y G, XU X P, et al. Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(10): 5962-5967. |
| [77] | ZHU Q L, LIU Y G. TransGene stacking Ⅱ vector system for plant metabolic engineering and synthetic biology[J]. Methods in Molecular Biology, 2021, 2238: 19-35. |
| [78] | SRIVASTAVA V, THOMSON J. Gene stacking by recombinases[J]. Plant Biotechnology Journal, 2016, 14(2): 471-482. |
| [79] | ZHAO Y C, HAN J L, TAN J T, et al. Efficient assembly of long DNA fragments and multiple genes with improved nickase-based cloning and Cre/loxP recombination[J]. Plant Biotechnology Journal, 2022, 20(10): 1983-1995. |
| [80] | ZHU Q L, ZENG D C, YU S Z, et al. From Golden Rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm[J]. Molecular Plant, 2018, 11(12): 1440-1448. |
| [81] | ZHU Q L, YU S Z, ZENG D C, et al. Development of “Purple Endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system[J]. Molecular Plant, 2017, 10(7): 918-929. |
| [82] | WANG L M, SHEN B R, LI B D, et al. A synthetic photorespiratory shortcut enhances photosynthesis to boost biomass and grain yield in rice[J]. Molecular Plant, 2020, 13(12): 1802-1815. |
| [83] | SUN C, LI H C, LIU Y J, et al. Iterative recombinase technologies for efficient and precise genome engineering across kilobase to megabase scales[J]. Cell, 2025, 188(17): 4693-4710.e15. |
| [84] | RENSING S A, GOFFINET B, MEYBERG R, et al. The moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants[J]. The Plant Cell, 2020, 32(5): 1361-1376. |
| [85] | KAMISUGI Y, CUMING A C, COVE D J. Parameters determining the efficiency of gene targeting in the moss Physcomitrella patens [J]. Nucleic Acids Research, 2005, 33(19): e173. |
| [86] | BEN-TOV D, MAFESSONI F, CUCUY A, et al. Uncovering the dynamics of precise repair at CRISPR/Cas9-induced double-strand breaks[J]. Nature Communications, 2024, 15: 5096. |
| [87] | JEONG S H, LEE H J, LEE S J. Recent advances in CRISPR-Cas technologies for synthetic biology[J]. Journal of Microbiology, 2023, 61(1): 13-36. |
| [13] | BOCK R. Transgenic plastids in basic research and plant biotechnology[J]. Journal of Molecular Biology, 2001, 312(3): 425-438. |
| [14] | TAUNT H N, STOFFELS L, PURTON S. Green biologics: the algal chloroplast as a platform for making biopharmaceuticals[J]. Bioengineered, 2018, 9(1): 48-54. |
| [15] | WANI S H, HAIDER N, KUMAR H, et al. Plant plastid engineering[J]. Current Genomics, 2010, 11(7): 500-512. |
| [16] | JAMES J S, DAI J B, CHEW W L, et al. The design and engineering of synthetic genomes[J]. Nature Reviews Genetics, 2025, 26(5): 298-319. |
| [17] | PUCHTA H, HOUBEN A. Plant chromosome engineering-past, present and future[J]. New Phytologist, 2024, 241(2): 541-552. |
| [18] | YU W C, HAN F P, GAO Z, et al. Construction and behavior of engineered minichromosomes in maize[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(21): 8924-8929. |
| [19] | ZHONG C X, MARSHALL J B, TOPP C, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3[J]. The Plant Cell, 2002, 14(11): 2825-2836. |
| [20] | GENT J I, WANG N, DAWE R K. Stable centromere positioning in diverse sequence contexts of complex and satellite centromeres of maize and wild relatives[J]. Genome Biology, 2017, 18(1): 121. |
| [21] | ZÁVODNÍK M, FAJKUS P, FRANEK M, et al. Telomerase RNA gene paralogs in plants - the usual pathway to unusual telomeres[J]. New Phytologist, 2023, 239(6): 2353-2366. |
| [22] | KUMAWAT S, CHOI J Y. No end in sight: mysteries of the telomeric variation in plants[J]. American Journal of Botany, 2023, 110(11): e16244. |
| [23] | GRAHAM N D, CODY J P, SWYERS N C, et al. Chapter Three - Engineered minichromosomes in plants: structure, function, and applications[M/OL]//JEON K W. International review of cell and molecular biology. Netherlands, Amsterdan: Academic Press, 2015: 63-119. (2015-06-17)[2025-09-01]. . |
| [24] | BIRCHLER J A, GRAHAM N D, SWYERS N C, et al. Plant minichromosomes[J]. Current Opinion in Biotechnology, 2016, 37: 135-142. |
| [25] | YIN X Z, ZHANG Y X, CHEN Y H, et al. Precise characterization and tracking of stably inherited artificial minichromosomes made by telomere-mediated chromosome truncation in Brassica napus [J]. Frontiers in Plant Science, 2021, 12: 743792. |
| [88] | ROZOV S M, PERMYAKOVA N V, DEINEKO E V. The problem of the low rates of CRISPR/Cas9-mediated knock-ins in plants: approaches and solutions[J]. International Journal of Molecular Sciences, 2019, 20(13): 3371. |
| [89] | LIANG Z, CHEN K L, LI T D, et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes[J]. Nature Communications, 2017, 8: 14261. |
| [90] | WOO J W, KIM J, KWON S I, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins[J]. Nature Biotechnology, 2015, 33(11): 1162-1164. |
| [91] | OSTROV N, BEAL J, ELLIS T, et al. Technological challenges and milestones for writing genomes[J]. Science, 2019, 366(6463): 310-312. |
| [92] | LIU W S, STEWART C N JR. Plant synthetic biology[J]. Trends in Plant Science, 2015, 20(5): 309-317. |
| [93] | VOLLEN K, ZHAO C S, ALONSO J M, et al. Sourcing DNA parts for synthetic biology applications in plants[J]. Current Opinion in Biotechnology, 2024, 87: 103140. |
| [94] | PFOTENHAUER A C, OCCHIALINI A, NGUYEN M A, et al. Building the plant SynBio toolbox through combinatorial analysis of DNA regulatory elements[J]. ACS Synthetic Biology, 2022, 11(8): 2741-2755. |
| [95] | NAYAK N, MEHROTRA S, KARAMCHANDANI A N, et al. Recent advances in designing synthetic plant regulatory modules[J]. Frontiers in Plant Science, 2025, 16: 1567659. |
| [96] | TIAN C F, LI J H, WU Y H, et al. An integrative database and its application for plant synthetic biology research[J]. Plant Communications, 2024, 5(5): 100827. |
| [97] | SILVEIRA GOMIDE M DA, DE CASTRO LEITÃO M, COELHO C M. Biocircuits in plants and eukaryotic algae[J]. Frontiers in Plant Science, 2022, 13: 982959. |
| [98] | BASSO M F, ARRAES F B M, GROSSI-DE-SA M, et al. Insights into genetic and molecular elements for transgenic crop development[J]. Frontiers in Plant Science, 2020, 11: 509. |
| [99] | ANDRES J, BLOMEIER T, ZURBRIGGEN M D. Synthetic switches and regulatory circuits in plants[J]. Plant Physiology, 2019, 179(3): 862-884. |
| [26] | GAETA R T, MASONBRINK R E, ZHAO C Z, et al. In vivo modification of a maize engineered minichromosome[J]. Chromosoma, 2013, 122(3): 221-232. |
| [27] | YAN X H, LI C, YANG J, et al. Induction of telomere-mediated chromosomal truncation and behavior of truncated chromosomes in Brassica napus [J]. The Plant Journal, 2017, 91(4): 700-713. |
| [28] | KAPUSI E, MA L, TEO C H, et al. Telomere-mediated truncation of barley chromosomes[J]. Chromosoma, 2012, 121(2): 181-190. |
| [29] | TEO C H, MA L, KAPUSI E, et al. Induction of telomere-mediated chromosomal truncation and stability of truncated chromosomes in Arabidopsis thaliana [J]. The Plant Journal, 2011, 68(1): 28-39. |
| [30] | NELSON A D, LAMB J C, KOBROSSLY P S, et al. Parameters affecting telomere-mediated chromosomal truncation in Arabidopsis [J]. The Plant Cell, 2011, 23(6): 2263-2272. |
| [31] | XU C H, CHENG Z K, YU W C. Construction of rice mini-chromosomes by telomere-mediated chromosomal truncation[J]. The Plant Journal, 2012, 70(6): 1070-1079. |
| [32] | YAN D, MA Y H, WANG H, et al. High ionic conductivity conjugated artificial solid electrolyte interphase enabling stable lithium metal batteries[J]. Green Chemistry, 2025, 27(25): 7564-7574. |
| [33] | LIU J L, ZHANG R X, CHAI N, et al. Programmable genome engineering and gene modifications for plant biodesign[J]. Plant Communications, 2025, 6(8): 101427. |
| [34] | YANG X Y, LI J H, CHEN L, et al. Stable mitotic inheritance of rice minichromosomes in cell suspension cultures[J]. Plant Cell Reports, 2015, 34(6): 929-941. |
| [35] | SATTAR M N, HASHEDI S A AL, MUNIR M, et al. Practical applications of minichromosomes in modern agriculture for better crops[M]//AL-KHAYRI J M, YATOO A M, JAIN S M, et al. Handbook of agricultural technologies. Singapore: Springer Nature Singapore, 2025: 1-22. (2025-02-28)[2025-09-01]. . |
| [36] | JONES N, HOUBEN A. B chromosomes in plants: escapees from the A chromosome genome [J]. Trends in Plant Science, 2003, 8(9): 417-423. |
| [37] | BIRCHLER J A, SWYERS N C. Engineered minichromosomes in plants[J]. Experimental Cell Research, 2020, 388(2): 111852. |
| [38] | NAISH M. Bridging the gap: unravelling plant centromeres in the telomere-to-telomere era[J]. New Phytologist, 2024, 244(6): 2143-2149. |
| [39] | COMAI L, MAHESHWARI S, MARIMUTHU M P A. Plant centromeres[J]. Current Opinion in Plant Biology, 2017, 36: 158-167. |
| [100] | EMILIANI V, ENTCHEVA E, HEDRICH R, et al. Optogenetics for light control of biological systems[J]. Nature Reviews Methods Primers, 2022, 2: 55. |
| [101] | KONG C, YANG Y, QI T C, et al. Predictive genetic circuit design for phenotype reprogramming in plants[J]. Nature Communications, 2025, 16: 715. |
| [102] | VAZQUEZ-VILAR M, QUIJANO-RUBIO A, FERNANDEZ-DEL-CARMEN A, et al. GB3.0: a platform for plant bio-design that connects functional DNA elements with associated biological data[J]. Nucleic Acids Research, 2017, 45(4): 2196-2209. |
| [103] | YOO S D, CHO Y H, SHEEN J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis[J]. Nature Protocols, 2007, 2(7): 1565-1572. |
| [104] | JIANG F W, ZHU J, LIU H L. Protoplasts: a useful research system for plant cell biology, especially dedifferentiation[J]. Protoplasma, 2013, 250(6): 1231-1238. |
| [105] | DLUGOSZ E M, LENAGHAN S C, STEWART C N. A robotic platform for high-throughput protoplast isolation and transformation[J]. Journal of Visualized Experiments, 2016(115): e54300. |
| [106] | ONG J Y, SWIDAH R, MONTI M, et al. SCRaMbLE: a study of its robustness and challenges through enhancement of hygromycin B resistance in a semi-synthetic yeast[J]. Bioengineering, 2021, 8(3): 42. |
| [107] | SCHINDLER D, WALKER R S K, JIANG S Y, et al. Design, construction, and functional characterization of a tRNA neochromosome in yeast[J]. Cell, 2023, 186(24): 5237-5253.e22. |
| [108] | NGUYEN E, POLI M, DURRANT M G, et al. Sequence modeling and design from molecular to genome scale with Evo[J]. Science, 2024, 386(6723): eado9336. |
| [109] | AVSEC Ž, LATYSHEVA N, CHENG J, et al. AlphaGenome: advancing regulatory variant effect prediction with a unified DNA sequence model[EB/OL]. bioRxiv. 2025. (2025-07-11)[2025-09-01]. . |
| [110] | PRADHAN B, PANDA D, BISHI S K, et al. Progress and prospects of C4 trait engineering in plants[J]. Plant Biology, 2022, 24(6): 920-931. |
| [111] | PRYWES N, PHILLIPS N R, TUCK O T, et al. Rubisco function, evolution, and engineering[J]. Annual Review of Biochemistry, 2023, 92: 385-410. |
| [40] | YU W C, YAU Y Y, BIRCHLER J A. Plant artificial chromosome technology and its potential application in genetic engineering[J]. Plant Biotechnology Journal, 2016, 14(5): 1175-1182. |
| [41] | ANANIEV E V, WU C C, CHAMBERLIN M A, et al. Artificial chromosome formation in maize (Zea mays L.)[J]. Chromosoma, 2009, 118(2): 157-177. |
| [42] | LOGSDON G A, GAMBOGI C W, LISKOVYKH M A, et al. Human artificial chromosomes that bypass centromeric DNA[J]. Cell, 2019, 178(3): 624-639.e19. |
| [43] | MENDIBURO M J, PADEKEN J, FÜLÖP S, et al. Drosophila CENH3 is sufficient for centromere formation[J]. Science, 2011, 334(6056): 686-690. |
| [44] | ZENG Y B, WANG M Y, GENT J I, et al. Increased maize chromosome number by engineered chromosome fission[J]. Science Advances, 2025, 11(21): eadw3433. |
| [45] | DAWE R K, GENT J I, ZENG Y B, et al. Synthetic maize centromeres transmit chromosomes across generations[J]. Nature Plants, 2023, 9(3): 433-441. |
| [46] | PESKA V, GARCIA S. Origin, diversity, and evolution of telomere sequences in plants[J]. Frontiers in Plant Science, 2020, 11: 117. |
| [47] | TAN X Y, WU X L, HAN M Z, et al. Yeast autonomously replicating sequence (ARS): identification, function, and modification[J]. Engineering in Life Sciences, 2021, 21(7): 464-474. |
| [48] | ECKDAHL T T, BENNETZEN J L, ANDERSON J N. DNA structures associated with autonomously replicating sequences from plants[J]. Plant Molecular Biology, 1989, 12(5): 507-516. |
| [49] | MOLIN W T, YAGUCHI A, BLENNER M, et al. Autonomous replication sequences from the Amaranthus palmeri eccDNA replicon enable replication in yeast[J]. BMC Research Notes, 2020, 13(1): 330. |
| [50] | OCCHIALINI A, PFOTENHAUER A C, LI L, et al. Mini-synplastomes for plastid genetic engineering[J]. Plant Biotechnology Journal, 2022, 20(2): 360-373. |
| [51] | OCCHIALINI A, KING G, MAJDI M, et al. An optimized version of the small synthetic genome (mini-synplastome) for plastid metabolic engineering in Solanum tuberosum (potato)[J]. ACS Synthetic Biology, 2024, 13(12): 4245-4257. |
| [52] | YONG J X, WU M M, CARROLL B J, et al. Enhancing plant biotechnology by nanoparticle delivery of nucleic acids[J]. Trends in Genetics, 2024, 40(4): 352-363. |
| [53] | RUSTGI S, NAVEED S, WINDHAM J, et al. Plant biomacromolecule delivery methods in the 21st century[J]. Frontiers in Genome Editing, 2022, 4: 1011934. |
| [54] | MIYAMOTO T, NUMATA K. Advancing biomolecule delivery in plants: harnessing synthetic nanocarriers to overcome multiscale barriers for cutting-edge plant bioengineering[J]. Bulletin of the Chemical Society of Japan, 2023, 96(9): 1026-1044. |
| [55] | CHO H J, MOY Y, RUDNICK N A, et al. Development of an efficient marker-free soybean transformation method using the novel bacterium Ochrobactrum haywardense H1[J]. Plant Biotechnology Journal, 2022, 20(5): 977-990. |
| [56] | WOO S S, JIANG J, GILL B S, et al. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor [J]. Nucleic Acids Research, 1994, 22(23): 4922-4931. |
| [57] | WANG G L, HOLSTEN T E, SONG W Y, et al. Construction of a rice bacterial artificial chromosome library and identification of clones linked to the Xa-21 disease resistance locus[J]. The Plant Journal, 1995, 7(3): 525-533. |
| [58] | HAMILTON C M, FRARY A, LEWIS C, et al. Stable transfer of intact high molecular weight DNA into plant chromosomes[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(18): 9975-9979. |
| [59] | ZIEMIENOWICZ A. Agrobacterium-mediated plant transformation: factors, applications and recent advances[J]. Biocatalysis and Agricultural Biotechnology, 2014, 3(4): 95-102. |
| [60] | LI X Y, YANG Q H, PENG L, et al. Agrobacterium-delivered VirE2 interacts with host nucleoporin CG1 to facilitate the nuclear import of VirE2-coated T complex[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(42): 26389-26397. |
| [61] | GORALOGIA G S, WILLIG C, STRAUSS S H. Engineering Agrobacterium for improved plant transformation[J]. The Plant Journal, 2025, 121(5): e70015. |
| [62] | LIU Y C, VIDALI L. Efficient polyethylene glycol (PEG) mediated transformation of the moss Physcomitrella patens [J]. Journal of Visualized Experiments, 2011(50): e2560. |
| [63] | BATES G W. Plant transformation via protoplast electroporation[M/OL]//HALL R D. Methods in molecular biology: plant cell culture protocols. Totowa, NJ: Humana Press, 1999: 359-366 [2025-09-01]. . |
| [64] | MORI K, TANASE K, SASAKI K. Novel electroporation-based genome editing of carnation plant tissues using RNPs targeting the anthocyanidin synthase gene[J]. Planta, 2024, 259(4): 84. |
| [65] | LEE K, WANG K. Strategies for genotype-flexible plant transformation[J]. Current Opinion in Biotechnology, 2023, 79: 102848. |
| [66] | ISMAGUL A, YANG N N, MALTSEVA E, et al. A biolistic method for high-throughput production of transgenic wheat plants with single gene insertions[J]. BMC Plant Biology, 2018, 18(1): 135. |
| [67] | KANDHOL N, DASH P K, SINGH V P, et al. Nanomaterial-based gene delivery in plants: an upcoming genetic revolution [J/OL]. Trends in Plant Science, 2025. (2025-06-28)[2025-09-01]. . |
| [68] | YAN Y, ZHU X J, YU Y, et al. Nanotechnology strategies for plant genetic engineering[J]. Advanced Materials, 2022, 34(7): 2106945. |
| [69] | ZUVIN M, KURUOGLU E, KAYA V O, et al. Magnetofection of green fluorescent protein encoding DNA-bearing polyethyleneimine-coated superparamagnetic iron oxide nanoparticles to human breast cancer cells[J]. ACS Omega, 2019, 4(7): 12366-12374. |
| [70] | LIU Y, YANG H, SAKANISHI A. Ultrasound: mechanical gene transfer into plant cells by sonoporation[J]. Biotechnology Advances, 2006, 24(1): 1-16. |
| [71] | YANG L Y, CUI G M, WANG Y X, et al. Expression of foreign genes demonstrates the effectiveness of pollen-mediated transformation in Zea mays [J]. Frontiers in Plant Science, 2017, 8: 383. |
| [72] | JOERSBO M, BRUNSTEDT J. Sonication: a new method for gene transfer to plants[J]. Physiologia Plantarum, 1992, 85(2): 230-234. |
| [73] | NANASATO Y, KONAGAYA K I, OKUZAKI A, et al. Agrobacterium-mediated transformation of kabocha squash (Cucurbita moschata Duch) induced by wounding with aluminum borate whiskers[J]. Plant Cell Reports, 2011, 30(8): 1455-1464. |
| [74] | MATSUSHITA J, OTANI M, WAKITA Y, et al. Transgenic plant regeneration through silicon carbide whisker-mediated transformation of rice (Oryza sativa L.)[J]. Breeding Science, 1999, 49(1): 21-26. |
| [112] | BAHUGUNA V, BHATT G, MAIKHURI R, et al. Nitrogen fixation through genetic engineering: a future systemic approach of nitrogen fixation[M/OL]//NATH M, BHATT D, BHARGAVA P, et al. Microbial metatranscriptomics belowground. Singapore: Springer Singapore, 2021: 109-122. (2021-06-03)[2025-09-01]. . |
| [113] | BENNETT E M, MURRAY J W, ISALAN M. Engineering nitrogenases for synthetic nitrogen fixation: from pathway engineering to directed evolution[J]. BioDesign Research, 2023, 5: 5. |
| [114] | MONTESINOS E. Functional peptides for plant disease control[J]. Annual Review of Phytopathology, 2023, 61: 301-324. |
| [115] | AHUJA I, KISSEN R, BONES A M. Phytoalexins in defense against pathogens[J]. Trends in Plant Science, 2012, 17(2): 73-90. |
| [116] | HUANG W K, ZHANG Y M, XIAO N, et al. Trans-complementation of the viral movement protein mediates efficient expression of large target genes via a tobacco mosaic virus vector[J]. Plant Biotechnology Journal, 2024, 22(11): 2957-2970. |
| [117] | HU Y J, GU C C, WANG X F, et al. Asymmetric total synthesis of taxol[J]. Journal of the American Chemical Society, 2021, 143(42): 17862-17870. |
| [118] | LI J H, MUTANDA I, WANG K B, et al. Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana [J]. Nature Communications, 2019, 10: 4850. |
| [119] | SU C, CUI H T, WANG W W, et al. Bioremediation of complex organic pollutants by engineered Vibrio natriegens [J]. Nature, 2025, 642(8069): 1024-1033. |
| [120] | BOEHM C R, BOCK R. Recent advances and current challenges in synthetic biology of the plastid genetic system and metabolism[J]. Plant Physiology, 2019, 179(3): 794-802. |
| [121] | ZHANG Y Z, YUAN J L, ZHANG L R, et al. Coupling of H3K27me3 recognition with transcriptional repression through the BAH-PHD-CPL2 complex in Arabidopsis [J]. Nature Communications, 2020, 11: 6212. |
| [1] | SONG Kainan, ZHANG Liwen, WANG Chao, TIAN Pingfang, LI Guangyue, PAN Guohui, XU Yuquan. Advances in small-molecule biopesticides and their biosynthesis [J]. Synthetic Biology Journal, 2025, 6(5): 1203-1223. |
| [2] | YU Wenwen, LV Xueqin, LI Zhaofeng, LIU Long. Plant synthetic biology and bioproduction of human milk oligosaccharides [J]. Synthetic Biology Journal, 2025, 6(5): 992-997. |
| [3] | YAN Zhaotao, ZHOU Pengfei, WANG Yangzhong, ZHANG Xin, XIE Wenyan, TIAN Chenfei, WANG Yong. Plant synthetic biology: new opportunities for large-scale culture of plant cells [J]. Synthetic Biology Journal, 2025, 6(5): 1107-1125. |
| [4] | SUN Yang, CHEN Lichao, SHI Yanyun, WANG Ke, LV Dandan, XU Xiumei, ZHANG Lixin. Strategies and prospects of synthetic biology in crop photosynthesis [J]. Synthetic Biology Journal, 2025, 6(5): 1025-1040. |
| [5] | ZHAO Xinyu, SHENG Qi, LIU Kaifang, LIU Jia, LIU Liming. Construction of microbial cell factories for aspartate-family feed amino acids [J]. Synthetic Biology Journal, 2025, 6(5): 1184-1202. |
| [6] | HE Yangyu, YANG Kai, WANG Weilin, HUANG Qian, QIU Ziying, SONG Tao, HE Liushang, YAO Jinxin, GAN Lu, HE Yuchi. Design and practice of plant synthetic biology theme in the International Genetically Engineered Machine Competition [J]. Synthetic Biology Journal, 2025, 6(5): 1243-1254. |
| [7] | ZHANG Xuebo, ZHU Chengshu, CHEN Ruiyun, JIN Qingzi, LIU Xiao, XIONG Yan, CHEN Daming. Policy planning and industrial development of agricultural synthetic biology [J]. Synthetic Biology Journal, 2025, 6(5): 1224-1242. |
| [8] | LIU Jie, GAO Yu, MA Yongshuo, SHANG Yi. Progress and challenges of synthetic biology in agriculture [J]. Synthetic Biology Journal, 2025, 6(5): 998-1024. |
| [9] | ZHENG Lei, ZHENG Qiteng, ZHANG Tianjiao, DUAN Kun, ZHANG Ruifu. Engineering rhizosphere synthetic microbial communities to enhance crop nutrient use efficiency [J]. Synthetic Biology Journal, 2025, 6(5): 1058-1071. |
| [10] | LI Chao, ZHANG Huan, YANG Jun, WANG Ertao. Research advances in nitrogen fixation synthetic biology [J]. Synthetic Biology Journal, 2025, 6(5): 1041-1057. |
| [11] | PU Ya, JIAO Yuling. Plant artificial chromosomes: current research progress and future application perspectives [J]. Synthetic Biology Journal, 2025, 6(5): 1072-1092. |
| [12] | FANG Xinyi, SUN Lichao, HUO Yixin, WANG Ying, YUE Haitao. Trends and challenges in microbial synthesis of higher alcohols [J]. Synthetic Biology Journal, 2025, 6(4): 873-898. |
| [13] | WU Xiaoyan, SONG Qi, XU Rui, DING Chenjun, CHEN Fang, GUO Qing, ZHANG Bo. A comparative analysis of global research and development competition in synthetic biology [J]. Synthetic Biology Journal, 2025, 6(4): 940-955. |
| [14] | ZHANG Jiankang, WANG Wenjun, GUO Hongju, BAI Beichen, ZHANG Yafei, YUAN Zheng, LI Yanhui, LI Hang. Development and application of a high-throughput microbial clone picking workstation based on machine vision [J]. Synthetic Biology Journal, 2025, 6(4): 956-971. |
| [15] | LI Quanfei, CHEN Qian, LIU Hao, HE Kundong, PAN Liang, LEI Peng, GU Yi’an, SUN Liang, LI Sha, QIU Yibin, WANG Rui, XU Hong. Synthetic biology and applications of high-adhesion protein materials [J]. Synthetic Biology Journal, 2025, 6(4): 806-828. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||