Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (5): 1058-1071.DOI: 10.12211/2096-8280.2025-075
• Invited Review • Previous Articles Next Articles
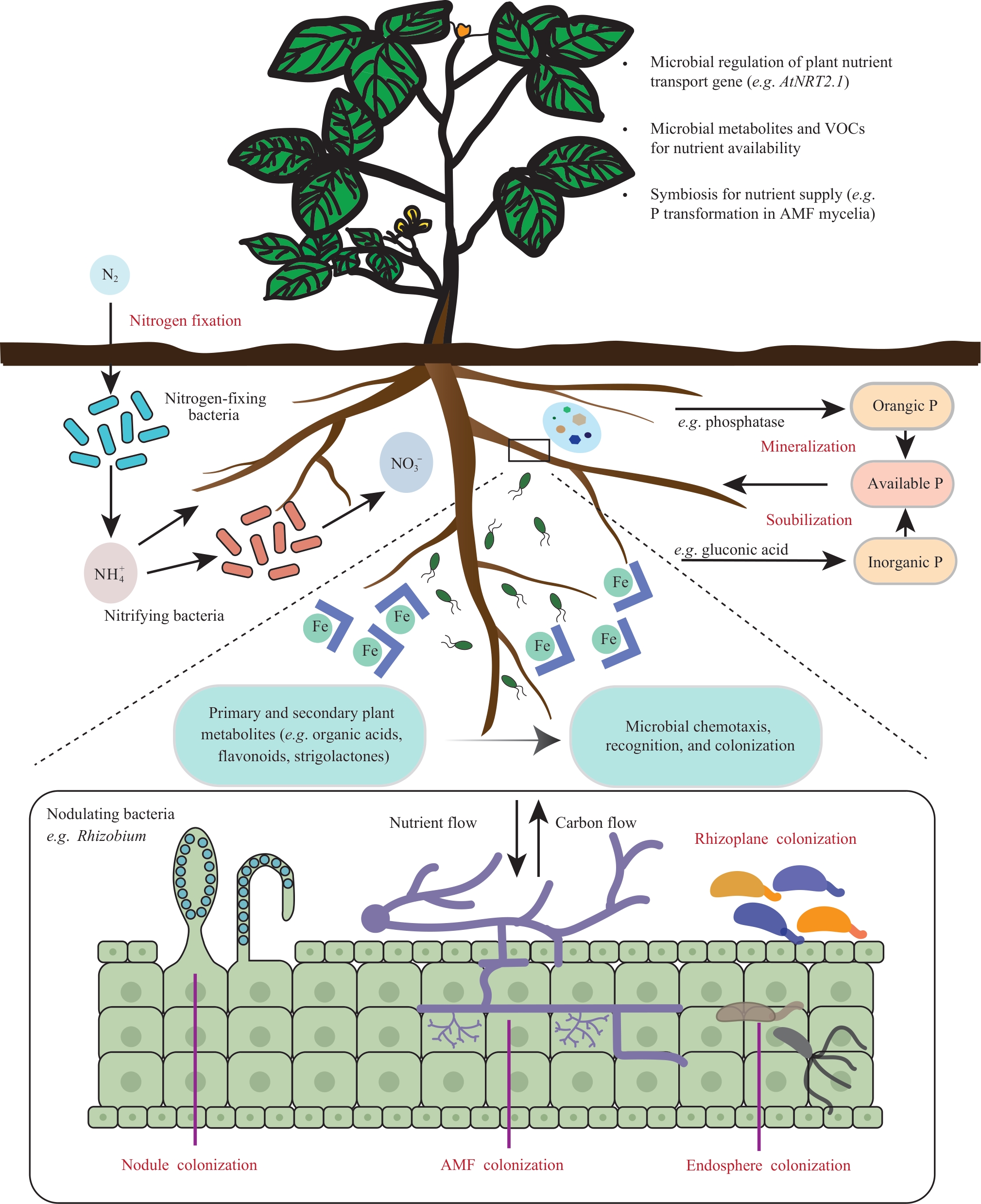
Engineering rhizosphere synthetic microbial communities to enhance crop nutrient use efficiency
ZHENG Lei, ZHENG Qiteng, ZHANG Tianjiao, DUAN Kun, ZHANG Ruifu
- The College of Resources and Environmental Sciences,Nanjing Agricultural University,Nanjing 210095,Jiangsu,China
-
Received:2025-07-21Revised:2025-09-09Online:2025-11-05Published:2025-10-31 -
Contact:ZHANG Ruifu
构建根际合成微生物菌群促进作物养分高效吸收利用
郑雷, 郑棋腾, 张天骄, 段鲲, 张瑞福
- 南京农业大学资源与环境科学学院,江苏 南京 210095
-
通讯作者:张瑞福 -
作者简介:郑雷 (1995—),男,博士研究生。研究方向为根际微生物功能解析与植物养分吸收调控。E-mail:zzzl@stu.njau.edu.cn张瑞福 (1974—),男,教授,博士生导师。研究方向为根际微生物与生物肥料、农业有机废弃物微生物降解转化与有机肥料等。E-mail:rfzhang@njau.edu.cn -
基金资助:国家自然科学基金国际(地区)合作与交流项目(32361143785)
CLC Number:
Cite this article
ZHENG Lei, ZHENG Qiteng, ZHANG Tianjiao, DUAN Kun, ZHANG Ruifu. Engineering rhizosphere synthetic microbial communities to enhance crop nutrient use efficiency[J]. Synthetic Biology Journal, 2025, 6(5): 1058-1071.
郑雷, 郑棋腾, 张天骄, 段鲲, 张瑞福. 构建根际合成微生物菌群促进作物养分高效吸收利用[J]. 合成生物学, 2025, 6(5): 1058-1071.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-075
| [1] | LAMBERS H, RAVEN J A, SHAVER G R, et al. Plant nutrient-acquisition strategies change with soil age[J]. Trends in Ecology & Evolution, 2008, 23(2): 95-103. |
| [2] | SHI J C, WANG X L, WANG E T. Mycorrhizal symbiosis in plant growth and stress adaptation: from genes to ecosystems[J]. Annual Review of Plant Biology, 2023, 74: 569-607. |
| [3] | YANG J, LAN L Y, JIN Y, et al. Mechanisms underlying legume-rhizobium symbioses[J]. Journal of Integrative Plant Biology, 2022, 64(2): 244-267. |
| [4] | PANG Z Q, CHEN J, WANG T H, et al. Linking plant secondary metabolites and plant microbiomes: a review[J]. Frontiers in Plant Science, 2021, 12: 621276. |
| [5] | VENTURI V, KEEL C. Signaling in the rhizosphere[J]. Trends in Plant Science, 2016, 21(3): 187-198. |
| [6] | ZHOU X G, ZHANG J Y, KHASHI U RAHMAN M, et al. Interspecific plant interaction via root exudates structures the disease suppressiveness of rhizosphere microbiomes[J]. Molecular Plant, 2023, 16(5): 849-864. |
| [7] | YU P, HE X M, BAER M, et al. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation[J]. Nature Plants, 2021, 7(4): 481-499. |
| [8] | SINGH B K, HU H W, MACDONALD C A, et al. Microbiome-facilitated plant nutrient acquisition[J]. Cell Host & Microbe, 2025, 33(6): 869-881. |
| [9] | TRIVEDI P, LEACH J E, TRINGE S G, et al. Plant-microbiome interactions: from community assembly to plant health[J]. Nature Reviews Microbiology, 2020, 18(11): 607-621. |
| [10] | GRIFFIN C, OZ M T, DEMIRER G S. Engineering plant-microbe communication for plant nutrient use efficiency[J]. Current Opinion in Biotechnology, 2024, 88: 103150. |
| [11] | DAI R, ZHANG J Y, LIU F, et al. Crop root bacterial and viral genomes reveal unexplored species and microbiome patterns[J]. Cell, 2025, 188(9): 2521-2539.e22. |
| [12] | DONG Q Q, SU H J, SUN Y X, et al. Metagenomic insights into nitrogen cycling functional gene responses to nitrogen fixation and transfer in maize-peanut intercropping[J]. Plant, Cell & Environment, 2024, 47(12): 4557-4571. |
| [13] | ELIAS M, TANAKA M, SAKAI M, et al. C-terminal periplasmic domain of Escherichia coli quinoprotein glucose dehydrogenase transfers electrons to ubiquinone[J]. Journal of Biological Chemistry, 2001, 276(51): 48356-48361. |
| [14] | LIU Y, JIA B L, REN Y, et al. Bacterial social interactions in synthetic Bacillus consortia enhance plant growth[J]. iMeta, 2025, 4(4): e70053. |
| [15] | BERG G, KUSSTATSCHER P, ABDELFATTAH A, et al. Microbiome modulation-toward a better understanding of plant microbiome response to microbial inoculants[J]. Frontiers in Microbiology, 2021, 12: 650610. |
| [16] | ERLACHER A, CARDINALE M, GROSCH R, et al. The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome[J]. Frontiers in Microbiology, 2014, 5: 175. |
| [17] | JIANG W Y, BIKARD D, COX D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nature Biotechnology, 2013, 31(3): 233-239. |
| [18] | JOHNS N I, BLAZEJEWSKI T, GOMES A L, et al. Principles for designing synthetic microbial communities[J]. Current Opinion in Microbiology, 2016, 31: 146-153. |
| [19] | STENUIT B, AGATHOS S N. Deciphering microbial community robustness through synthetic ecology and molecular systems synecology[J]. Current Opinion in Biotechnology, 2015, 33: 305-317. |
| [20] | HU J, WEI Z, FRIMAN V P, et al. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression[J]. mBio, 2016, 7(6): e01790-16. |
| [21] | WANG J J, LI R C, ZHANG H, et al. Beneficial bacteria activate nutrients and promote wheat growth under conditions of reduced fertilizer application[J]. BMC Microbiology, 2020, 20(1): 38. |
| [22] | WANG J, UWIRAGIYE Y, CAO M M, et al. Global land use change impacts on soil nitrogen availability and environmental losses[J]. Environmental Science & Technology, 2025, 59(33): 17595-17605. |
| [23] | HINSINGER P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review[J]. Plant and Soil, 2001, 237(2): 173-195. |
| [24] | GUERINOT M L, YI Y. Iron: nutritious, noxious, and not readily available[J]. Plant Physiology, 1994, 104(3): 815-820. |
| [25] | GALLOWAY J N, TOWNSEND A R, ERISMAN J W, et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions[J]. Science, 2008, 320(5878): 889-892. |
| [26] | YUE H, YUE W J, JIAO S, et al. Plant domestication shapes rhizosphere microbiome assembly and metabolic functions[J]. Microbiome, 2023, 11(1): 70. |
| [27] | XU H R, LIU W D, HE Y H, et al. Plant-root microbiota interactions in nutrient utilization[J]. Frontiers of Agricultural Science and Engineering, 2025, 12(1): 16-26. |
| [28] | CRAINE J M, MORROW C, FIERER N. Microbial nitrogen limitation increases decomposition[J]. Ecology, 2007, 88(8): 2105-2113. |
| [29] | FONTAINE S, HENAULT C, AAMOR A, et al. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect[J]. Soil Biology and Biochemistry, 2011, 43(1): 86-96. |
| [30] | DAKORA F D, PHILLIPS D A. Root exudates as mediators of mineral acquisition in low-nutrient environments[J]. Plant and Soil, 2002, 245(1): 35-47. |
| [31] | LIU S B, HE F K, KUZYAKOV Y, et al. Nutrients in the rhizosphere: a meta-analysis of content, availability, and influencing factors[J]. Science of the Total Environment, 2022, 826: 153908. |
| [32] | SCHALK I J. Bacterial siderophores: diversity, uptake pathways and applications[J]. Nature Reviews Microbiology, 2025, 23(1): 24-40. |
| [33] | FAN X Y, GE A H, QI S S, et al. Root exudates and microbial metabolites: signals and nutrients in plant-microbe interactions[J]. Science China Life Sciences, 2025, 68(8): 2290-2302. |
| [34] | KUYPERS M M M, MARCHANT H K, KARTAL B. The microbial nitrogen-cycling network[J]. Nature Reviews Microbiology, 2018, 16(5): 263-276. |
| [35] | CHENG S S, GONG X, XUE W F, et al. Evolutionarily conserved core microbiota as an extended trait in nitrogen acquisition strategy of herbaceous species[J]. New Phytologist, 2024, 244(4): 1570-1584. |
| [36] | YANG N, NESME J, RØDER H L, et al. Emergent bacterial community properties induce enhanced drought tolerance in Arabidopsis [J]. NPJ Biofilms and Microbiomes, 2021, 7: 82. |
| [37] | PORTER S S, DUPIN S E, DENISON R F, et al. Host-imposed control mechanisms in legume-rhizobia symbiosis[J]. Nature Microbiology, 2024, 9(8): 1929-1939. |
| [38] | PRIYA H, DHAR D W, SINGH R, et al. Co-cultivation approach to decipher the influence of nitrogen-fixing Cyanobacterium on growth and N uptake in rice crop[J]. Current Microbiology, 2022, 79(2): 53. |
| [39] | HUREK T, REINHOLD-HUREK B. Azoarcus sp. strain BH72 as a model for nitrogen-fixing grass endophytes[J]. Journal of Biotechnology, 2003, 106(2-3): 169-178. |
| [40] | WALLER S, WILDER S L, SCHUELLER M J, et al. Examining the effects of the nitrogen environment on growth and N2-fixation of endophytic Herbaspirillum seropedicae in maize seedlings by applying 11C radiotracing[J]. Microorganisms, 2021, 9(8): 1582. |
| [41] | STEENHOUDT O, VANDERLEYDEN J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects[J]. FEMS Microbiology Reviews, 2000, 24(4): 487-506. |
| [42] | HASKETT T L, PARAMASIVAN P, MENDES M D, et al. Engineered plant control of associative nitrogen fixation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(16): e2117465119. |
| [43] | ZHANG L Y, ZHANG M L, HUANG S Y, et al. A highly conserved core bacterial microbiota with nitrogen-fixation capacity inhabits the xylem sap in maize plants[J]. Nature Communications, 2022, 13: 3361. |
| [44] | VAN DEYNZE A, ZAMORA P, DELAUX P M, et al. Nitrogen fixation in a Landrace of maize is supported by a mucilage-associated diazotrophic microbiota[J]. PLoS Biology, 2018, 16(8): e2006352. |
| [45] | CALVO P, ZEBELO S, MCNEAR D, et al. Plant growth-promoting rhizobacteria induce changes in Arabidopsis thaliana gene expression of nitrate and ammonium uptake genes[J]. Journal of Plant Interactions, 2019, 14(1): 224-231. |
| [46] | CHEN Y, LI Y C, FU Y S, et al. The beneficial rhizobacterium Bacillus velezensis SQR9 regulates plant nitrogen uptake via an endogenous signaling pathway[J]. Journal of Experimental Botany, 2024, 75(11): 3388-3400. |
| [47] | MCGRATH J W, CHIN J P, QUINN J P. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules[J]. Nature Reviews Microbiology, 2013, 11(6): 412-419. |
| [48] | GROSS A, LIN Y, WEBER P K, et al. The role of soil redox conditions in microbial phosphorus cycling in humid tropical forests[J]. Ecology, 2020, 101(2): e02928. |
| [49] | LIANG J L, LIU J, JIA P, et al. Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining[J]. The ISME Journal, 2020, 14(6): 1600-1613. |
| [50] | BILLAH M, KHAN M, BANO A, et al. Phosphorus and phosphate solubilizing bacteria: keys for sustainable agriculture[J]. Geomicrobiology Journal, 2019, 36(10): 904-916. |
| [51] | ZHAO B Y, JIA X Q, YU N, et al. Microbe-dependent and independent nitrogen and phosphate acquisition and regulation in plants[J]. New Phytologist, 2024, 242(4): 1507-1522. |
| [52] | DE ZUTTER N, AMEYE M, VERMEIR P, et al. Innovative rhizosphere-based enrichment under P-limitation selects for bacterial isolates with high-performance P-solubilizing traits[J]. Microbiology Spectrum, 2022, 10(6): e02052-22. |
| [53] | SINGH S K, WU X X, SHAO C Y, et al. Microbial enhancement of plant nutrient acquisition[J]. Stress Biology, 2022, 2(1): 3. |
| [54] | SHAO J H, MIAO Y Z, LIU K M, et al. Rhizosphere microbiome assembly involves seed-borne bacteria in compensatory phosphate solubilization[J]. Soil Biology and Biochemistry, 2021, 159: 108273. |
| [55] | PANG F, LI Q, SOLANKI M K, et al. Soil phosphorus transformation and plant uptake driven by phosphate-solubilizing microorganisms[J]. Frontiers in Microbiology, 2024, 15: 1383813. |
| [56] | LIU J P, XU W F, ZHANG Q, et al. OsPHR2-mediated recruitment of Pseudomonadaceae enhances rice phosphorus uptake[J]. Plant Communications, 2024, 5(8): 100930. |
| [57] | LIU C, BAI Z, LUO Y, et al. Multiomics dissection of Brassica napus L. lateral roots and endophytes interactions under phosphorus starvation[J]. Nature Communications, 2024, 15(1): 9732. |
| [58] | HARBORT C J, HASHIMOTO M, INOUE H, et al. Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis [J]. Cell Host & Microbe, 2020, 28(6): 825-837.e6. |
| [59] | WANG N Q, WANG T Q, CHEN Y, et al. Microbiome convergence enables siderophore-secreting-rhizobacteria to improve iron nutrition and yield of peanut intercropped with maize[J]. Nature Communications, 2024, 15: 839. |
| [60] | ZAMIOUDIS C, KORTELAND J, VAN PELT J A, et al. Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses[J]. The Plant Journal, 2015, 84(2): 309-322. |
| [61] | PRITY S A, SAJIB S A, DAS U, et al. Arbuscular mycorrhizal fungi mitigate Fe deficiency symptoms in sorghum through phytosiderophore-mediated Fe mobilization and restoration of redox status[J]. Protoplasma, 2020, 257(5): 1373-1385. |
| [62] | ZHANG P F, JIN T, KUMAR SAHU S, et al. The distribution of tryptophan-dependent indole-3-acetic acid synthesis pathways in bacteria unraveled by large-scale genomic analysis[J]. Molecules, 2019, 24(7): 1411. |
| [63] | NETT R S, MONTANARES M, MARCASSA A, et al. Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution[J]. Nature Chemical Biology, 2017, 13(1): 69-74. |
| [64] | SALAZAR-CEREZO S, MARTÍNEZ-MONTIEL N, GARCÍA-SÁNCHEZ J, et al. Gibberellin biosynthesis and metabolism: a convergent route for plants, fungi and bacteria[J]. Microbiological Research, 2018, 208: 85-98. |
| [65] | HAYASHI S, GRESSHOFF P M, FERGUSON B J. Mechanistic action of gibberellins in legume nodulation[J]. Journal of Integrative Plant Biology, 2014, 56(10): 971-978. |
| [66] | GROßKINSKY D K, TAFNER R, MORENO M V, et al. Cytokinin production by Pseudomonas fluorescens G20-18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis [J]. Scientific Reports, 2016, 6: 23310. |
| [67] | NUMAN M, BASHIR S, KHAN Y, et al. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review[J]. Microbiological Research, 2018, 209: 21-32. |
| [68] | CONWAY J M, WALTON W G, SALAS-GONZÁLEZ I, et al. Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome[J]. Nature Microbiology, 2022, 7(11): 1817-1833. |
| [69] | PACESA M, PELEA O, JINEK M. Past, present, and future of CRISPR genome editing technologies[J]. Cell, 2024, 187(5): 1076-1100. |
| [70] | CHEN L, ZHANG S, XUE N N, et al. Engineering a precise adenine base editor with minimal bystander editing[J]. Nature Chemical Biology, 2023, 19(1): 101-110. |
| [71] | XU P Y, SAITO M, FAURE G, et al. Structural insights into the diversity and DNA cleavage mechanism of Fanzor[J]. Cell, 2024, 187(19): 5238-5252.e20. |
| [72] | WEI Y H, GAO P F, PAN D, et al. Engineering eukaryotic transposon-encoded Fanzor2 system for genome editing in mammals[J]. Nature Chemical Biology, 2025. (2025-05-20)[2025-09-01]. . |
| [73] | HUA Y, TAY N E S, YE X J, et al. Protein editing using a coordinated transposition reaction[J]. Science, 2025, 388(6742): 68-74. |
| [74] | DIXON R A, POSTGATE J R. Genetic transfer of nitrogen fixation from Klebsiella pneumoniae to Escherichia coli [J]. Nature, 1972, 237(5350): 102-103. |
| [75] | TEMME K, ZHAO D H, VOIGT C A. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca [J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(18): 7085-7090. |
| [76] | WANG L Y, ZHANG L H, LIU Z Z, et al. A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli [J]. PLoS Genetics, 2013, 9(10): e1003865. |
| [77] | LI X X, LIU Q, LIU X M, et al. Using synthetic biology to increase nitrogenase activity[J]. Microbial Cell Factories, 2016, 15: 43. |
| [78] | ZHANG T, YAN Y L, HE S, et al. Involvement of the ammonium transporter AmtB in nitrogenase regulation and ammonium excretion in Pseudomonas stutzeri A1501[J]. Research in Microbiology, 2012, 163(5): 332-339. |
| [79] | BAGESHWAR U K, SRIVASTAVA M, PARDHA-SARADHI P, et al. An environmentally friendly engineered Azotobacter strain that replaces a substantial amount of urea fertilizer while sustaining the same wheat yield[J]. Applied and Environmental Microbiology, 2017, 83(15): e00590-17. |
| [80] | COMPANT S, CASSAN F, KOSTIĆ T, et al. Harnessing the plant microbiome for sustainable crop production[J]. Nature Reviews Microbiology, 2025, 23(1): 9-23. |
| [81] | WEN A, HAVENS K L, BLOCH S E, et al. Enabling biological nitrogen fixation for cereal crops in fertilized fields[J]. ACS Synthetic Biology, 2021, 10(12): 3264-3277. |
| [82] | RYU M H, ZHANG J, TOTH T, et al. Control of nitrogen fixation in bacteria that associate with cereals[J]. Nature Microbiology, 2020, 5(2): 314-330. |
| [83] | ALLEN R S, TILBROOK K, WARDEN A C, et al. Expression of 16 nitrogenase proteins within the plant mitochondrial matrix[J]. Frontiers in Plant Science, 2017, 8: 287. |
| [84] | HE W S, BURÉN S, BAYSAL C, et al. Nitrogenase cofactor maturase NifB isolated from transgenic rice is active in FeMo-co synthesis[J]. ACS Synthetic Biology, 2022, 11(9): 3028-3036. |
| [85] | ALORI E T, GLICK B R, BABALOLA O O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture[J]. Frontiers in Microbiology, 2017, 8: 971. |
| [86] | BUCH A, ARCHANA G, NARESH KUMAR G. Heterologous expression of phosphoenolpyruvate carboxylase enhances the phosphate solubilizing ability of fluorescent pseudomonads by altering the glucose catabolism to improve biomass yield[J]. Bioresource Technology, 2010, 101(2): 679-687. |
| [87] | ADHIKARY H, SANGHAVI P B, MACWAN S R, et al. Artificial citrate operon confers mineral phosphate solubilization ability to diverse fluorescent pseudomonads[J]. PLoS One, 2014, 9(9): e107554. |
| [88] | SHULSE C N, CHOVATIA M, AGOSTO C, et al. Engineered root bacteria release plant-available phosphate from phytate[J]. Applied and Environmental Microbiology, 2019, 85(18): e01210-19. |
| [89] | LIU Y P, SHU X, CHEN L, et al. Plant commensal type Ⅶ secretion system causes iron leakage from roots to promote colonization[J]. Nature Microbiology, 2023, 8(8): 1434-1449. |
| [90] | CHEN J W, ZHANG X, KUANG M, et al. Endophytic Enterobacter sp. YG-14 mediated arsenic mobilization through siderophore and its role in enhancing phytostabilization[J]. Journal of Hazardous Materials, 2024, 465: 133206. |
| [91] | DE SOUZA R S C, ARMANHI J S L, ARRUDA P. From microbiome to traits: designing synthetic microbial communities for improved crop resiliency[J]. Frontiers in Plant Science, 2020, 11: 1179. |
| [92] | XI H C, NIE X Q, GAO F, et al. A bacterial spermidine biosynthetic pathway via carboxyaminopropylagmatine[J]. Science Advances, 2023, 9(43): eadj9075. |
| [93] | SHEN J Y, WANG M X, WANG E T. Exploitation of the microbiome for crop breeding[J]. Nature Plants, 2024, 10(4): 533-534. |
| [94] | VORHOLT J A, VOGEL C, CARLSTRÖM C I, et al. Establishing causality: opportunities of synthetic communities for plant microbiome research[J]. Cell Host & Microbe, 2017, 22(2): 142-155. |
| [95] | JING J Y, GARBEVA P, RAAIJMAKERS J M, et al. Strategies for tailoring functional microbial synthetic communities[J]. The ISME Journal, 2024, 18(1): wrae049. |
| [96] | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes[J]. Nature Reviews Microbiology, 2019, 17(12): 725-741. |
| [97] | WANG W, XIA Y W, ZHANG P P, et al. Narrow-spectrum resource-utilizing bacteria drive the stability of synthetic communities through enhancing metabolic interactions[J]. Nature Communications, 2025, 16: 6088. |
| [98] | KAUR S, EGIDI E, QIU Z G, et al. Synthetic community improves crop performance and alters rhizosphere microbial communities[J]. Journal of Sustainable Agriculture and Environment, 2022, 1(2): 118-131. |
| [99] | CHAI Y N, GE Y F, STOERGER V, et al. High-resolution phenotyping of sorghum genotypic and phenotypic responses to low nitrogen and synthetic microbial communities[J]. Plant, Cell & Environment, 2021, 44(5): 1611-1626. |
| [100] | NIU B, PAULSON J N, ZHENG X Q, et al. Simplified and representative bacterial community of maize roots[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(12): E2450-E2459. |
| [101] | XUN W B, REN Y, YAN H, et al. Sustained inhibition of maize seed-borne Fusarium using a Bacillus-dominated rhizospheric stable core microbiota with unique cooperative patterns[J]. Advanced Science, 2023, 10(5): 2205215. |
| [102] | QIAO Y Z, WANG Z D, SUN H, et al. Synthetic community derived from grafted watermelon rhizosphere provides protection for ungrafted watermelon against Fusarium oxysporum via microbial synergistic effects[J]. Microbiome, 2024, 12(1): 101. |
| [103] | XU X M, DINESEN C, PIOPPI A, et al. Composing a microbial symphony: synthetic communities for promoting plant growth[J]. Trends in Microbiology, 2025, 33(7): 738-751. |
| [104] | NORTHEN T R, KLEINER M, TORRES M, et al. Community standards and future opportunities for synthetic communities in plant-microbiota research[J]. Nature Microbiology, 2024, 9(11): 2774-2784. |
| [105] | WANG C H, LI Y J, LI M J, et al. Functional assembly of root-associated microbial consortia improves nutrient efficiency and yield in soybean[J]. Journal of Integrative Plant Biology, 2021, 63(6): 1021-1035. |
| [106] | LI Y J, LI R R, LIU R, et al. A simplified SynCom based on core-helper strain interactions enhances symbiotic nitrogen fixation in soybean[J]. Journal of Integrative Plant Biology, 2025, 67(6): 1582-1598. |
| [107] | LIU C Y, JIANG M T, YUAN M M, et al. Root microbiota confers rice resistance to aluminium toxicity and phosphorus deficiency in acidic soils[J]. Nature Food, 2023, 4(10): 912-924. |
| [108] | CASTRILLO G, TEIXEIRA P J P L, PAREDES S H, et al. Root microbiota drive direct integration of phosphate stress and immunity[J]. Nature, 2017, 543(7646): 513-518. |
| [109] | HASKETT T L, TKACZ A, POOLE P S. Engineering rhizobacteria for sustainable agriculture[J]. The ISME Journal, 2021, 15(4): 949-964. |
| [110] | OFEK M, VORONOV-GOLDMAN M, HADAR Y, et al. Host signature effect on plant root-associated microbiomes revealed through analyses of resident vs. active communities[J]. Environmental Microbiology, 2014, 16(7): 2157-2167. |
| [111] | ZHONG X B, WANG J, SHI X L, et al. Genetically optimizing soybean nodulation improves yield and protein content[J]. Nature Plants, 2024, 10(5): 736-742. |
| [1] | SONG Kainan, ZHANG Liwen, WANG Chao, TIAN Pingfang, LI Guangyue, PAN Guohui, XU Yuquan. Advances in small-molecule biopesticides and their biosynthesis [J]. Synthetic Biology Journal, 2025, 6(5): 1203-1223. |
| [2] | YU Wenwen, LV Xueqin, LI Zhaofeng, LIU Long. Plant synthetic biology and bioproduction of human milk oligosaccharides [J]. Synthetic Biology Journal, 2025, 6(5): 992-997. |
| [3] | YAN Zhaotao, ZHOU Pengfei, WANG Yangzhong, ZHANG Xin, XIE Wenyan, TIAN Chenfei, WANG Yong. Plant synthetic biology: new opportunities for large-scale culture of plant cells [J]. Synthetic Biology Journal, 2025, 6(5): 1107-1125. |
| [4] | SUN Yang, CHEN Lichao, SHI Yanyun, WANG Ke, LV Dandan, XU Xiumei, ZHANG Lixin. Strategies and prospects of synthetic biology in crop photosynthesis [J]. Synthetic Biology Journal, 2025, 6(5): 1025-1040. |
| [5] | ZHAO Xinyu, SHENG Qi, LIU Kaifang, LIU Jia, LIU Liming. Construction of microbial cell factories for aspartate-family feed amino acids [J]. Synthetic Biology Journal, 2025, 6(5): 1184-1202. |
| [6] | HE Yangyu, YANG Kai, WANG Weilin, HUANG Qian, QIU Ziying, SONG Tao, HE Liushang, YAO Jinxin, GAN Lu, HE Yuchi. Design and practice of plant synthetic biology theme in the International Genetically Engineered Machine Competition [J]. Synthetic Biology Journal, 2025, 6(5): 1243-1254. |
| [7] | ZHANG Xuebo, ZHU Chengshu, CHEN Ruiyun, JIN Qingzi, LIU Xiao, XIONG Yan, CHEN Daming. Policy planning and industrial development of agricultural synthetic biology [J]. Synthetic Biology Journal, 2025, 6(5): 1224-1242. |
| [8] | LIU Jie, GAO Yu, MA Yongshuo, SHANG Yi. Progress and challenges of synthetic biology in agriculture [J]. Synthetic Biology Journal, 2025, 6(5): 998-1024. |
| [9] | LI Chao, ZHANG Huan, YANG Jun, WANG Ertao. Research advances in nitrogen fixation synthetic biology [J]. Synthetic Biology Journal, 2025, 6(5): 1041-1057. |
| [10] | WEI Jiaxiu, JI Peiyun, JIE Qingyu, HUANG Qiuyan, YE Hao, DAI Junbiao. Construction and application of plant artificial chromosomes [J]. Synthetic Biology Journal, 2025, 6(5): 1093-1106. |
| [11] | FANG Xinyi, SUN Lichao, HUO Yixin, WANG Ying, YUE Haitao. Trends and challenges in microbial synthesis of higher alcohols [J]. Synthetic Biology Journal, 2025, 6(4): 873-898. |
| [12] | WU Xiaoyan, SONG Qi, XU Rui, DING Chenjun, CHEN Fang, GUO Qing, ZHANG Bo. A comparative analysis of global research and development competition in synthetic biology [J]. Synthetic Biology Journal, 2025, 6(4): 940-955. |
| [13] | ZHANG Jiankang, WANG Wenjun, GUO Hongju, BAI Beichen, ZHANG Yafei, YUAN Zheng, LI Yanhui, LI Hang. Development and application of a high-throughput microbial clone picking workstation based on machine vision [J]. Synthetic Biology Journal, 2025, 6(4): 956-971. |
| [14] | LI Quanfei, CHEN Qian, LIU Hao, HE Kundong, PAN Liang, LEI Peng, GU Yi’an, SUN Liang, LI Sha, QIU Yibin, WANG Rui, XU Hong. Synthetic biology and applications of high-adhesion protein materials [J]. Synthetic Biology Journal, 2025, 6(4): 806-828. |
| [15] | WU Ke, LUO Jiahao, LI Feiran. Applications of machine learning in the reconstruction and curation of genome-scale metabolic models [J]. Synthetic Biology Journal, 2025, 6(3): 566-584. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||