Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (1): 33-45.DOI: 10.12211/2096-8280.2020-064
• Invited Review • Previous Articles Next Articles
Enzymatic ligation technologies for the synthesis of pharmaceutical peptides and proteins
YANG Xinyu1,2, ZHU Tong1,2, LI Ruifeng1,2, WU Bian1
- 1.CAS Key Laboratory of Microbial Physiological and Metabolic Engineering,State Key Laboratory of Microbial Resources,Institute of Microbiology,Chinese Academy of Sciences,Beijing 100101,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2020-05-18Revised:2021-01-06Online:2021-03-12Published:2021-02-28 -
Contact:WU Bian
从药物多肽到蛋白质全合成:酶促拼接的方法原理与前沿应用
杨新宇1,2, 朱彤1,2, 李瑞峰1,2, 吴边1
- 1.中国科学院微生物研究所,中国科学院微生物生理与代谢工程重点实验室,微生物资源前期开发国家重点实验室,北京 100101
2.中国科学院大学生命科学学院,北京 100049
-
通讯作者:吴边 -
作者简介:杨新宇(1994—),男,硕士研究生,研究方向为酶促蛋白质化学合成与修饰。E-mail:yangxy@im.ac.cn
吴边(1982—),男,博士生导师,研究员,研究方向为微生物催化元件的深度挖掘、机理解析、合成设计。致力于将蛋白质计算机设计前沿方法引入酶工程的研究,改造复杂生物大分子结构,优化催化元件性能,并在此基础上构建重要药物前体的生物合成途径。E-mail:wub@im.ac.cn -
基金资助:国家重点研发计划(2018YFA0901600);国家自然科学基金优秀青年科学基金(31822002);国家自然科学基金面上项目(31870055)
CLC Number:
Cite this article
YANG Xinyu, ZHU Tong, LI Ruifeng, WU Bian. Enzymatic ligation technologies for the synthesis of pharmaceutical peptides and proteins[J]. Synthetic Biology Journal, 2021, 2(1): 33-45.
杨新宇, 朱彤, 李瑞峰, 吴边. 从药物多肽到蛋白质全合成:酶促拼接的方法原理与前沿应用[J]. 合成生物学, 2021, 2(1): 33-45.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-064
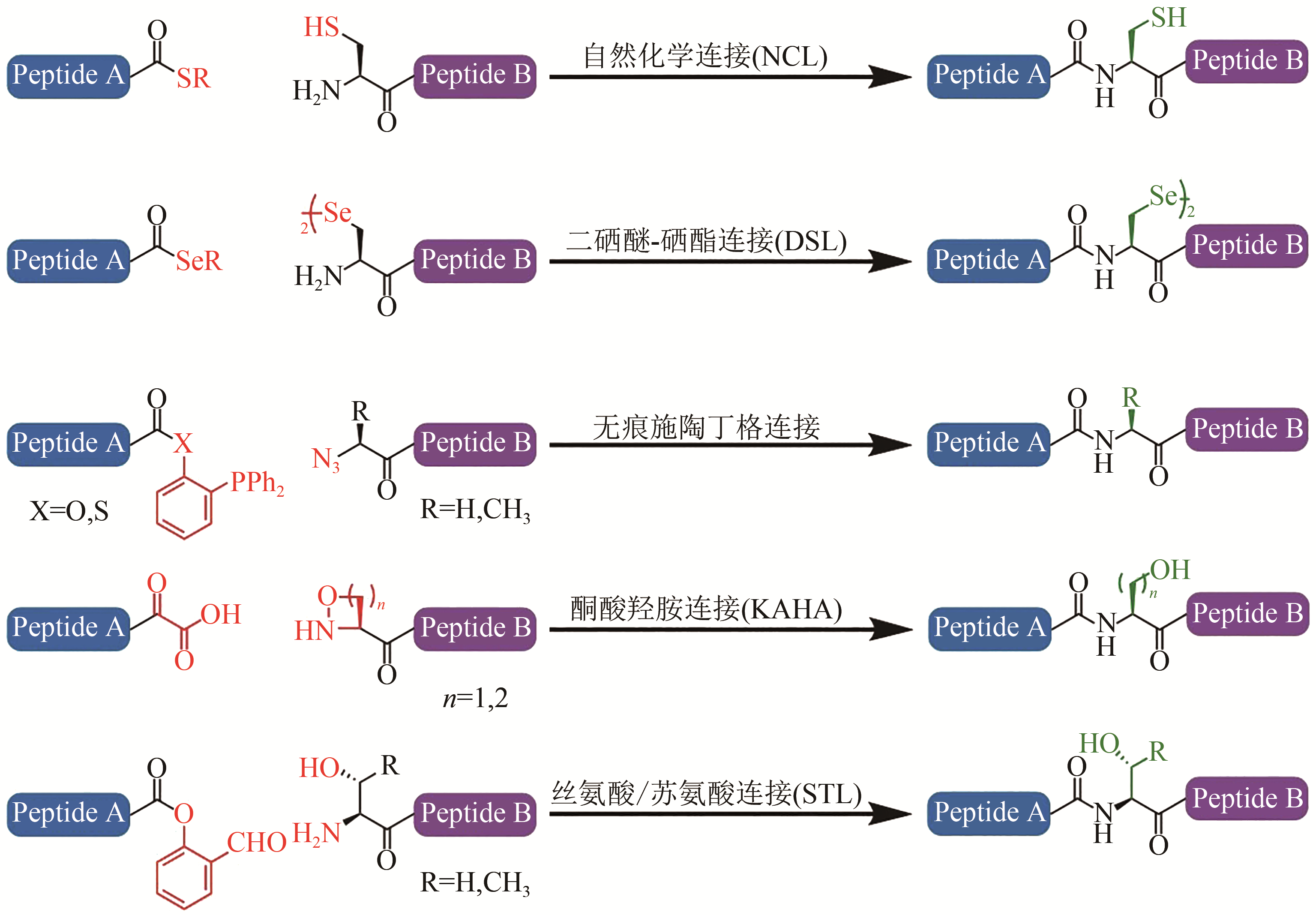
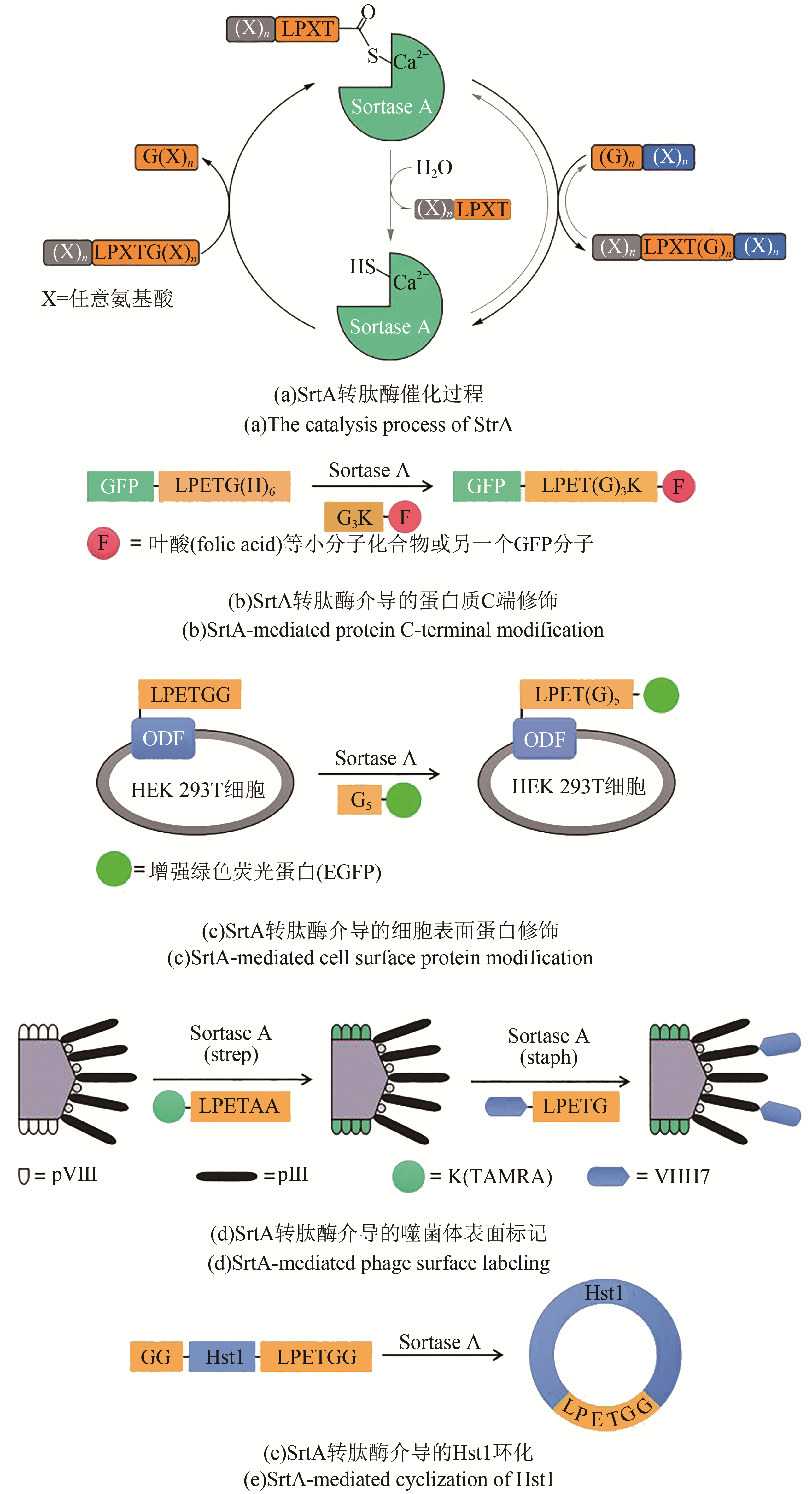
Fig. 1 Chemical ligation methods[13-17](Red groups participate the chemoselective capture reactions, and green groups represent ‘scar’ or sequence restrictions)
| 项目 | SrtA转肽酶 | Butelase 1转肽酶 | Subtilisin人工连接酶 | NCL | |
|---|---|---|---|---|---|
| 最初来源物种 | 金黄色葡萄球菌Staphylococcus aureus | 蝶豆 Clitoria ternatea | 解淀粉芽孢杆菌 Bacillus amyloliquefaciens | —— | |
| 酶的获取与产量[ | SrtA转肽酶可通过商业渠道获得或大肠杆菌重组表达 >40 mg/L | Butelase 1转肽酶通过植物材料提取 约5 mg/kg | Omniligase-1可通过商业渠道获得或枯草芽孢杆菌重组表达 >500 mg/L | —— | |
| 底物活化 | 底物无需活化 | 底物无需活化 | 多肽C端活化为 氧酯或硫酯 | 多肽C端活化为 硫酯 | |
连接 位点 序列 | C端 | LPXT-G | N/D-HV | X4X3X2X1—,X2外均避免Pro,X4偏好疏水&芳香族氨基酸 | 无限制 |
| N端 | (G) n | —X1X2,其中X1避免Pro和 酸性氨基酸,X2一般为ILVC | —X1’X2’均避免Pro | Cys | |
| 连接“疤痕” | LPXT(G) n | 一个Asn/Asp残基 | 无痕(X4X3X2X1 X1’X2’) | 一个Cys残基 | |
应用 范围 | 多肽首 尾环化 | 短肽(大于19个氨基酸残基)和蛋白质皆可环化 | 短肽(大于9个氨基酸残基)和蛋白质皆可环化 | 环化多肽一般不超过40个残基 | 环化多肽一般不超过40个残基 |
蛋白末 端修饰 | 蛋白质N端与C端皆可 | 蛋白质N端与C端皆可 | 大多用于蛋白质N端修饰鲜有C端修饰 | 蛋白质N端与C端皆可 | |
蛋白质 全合成 | 尚无应用报道 | 尚无应用报道 | 可以应用 | 广泛应用 | |
Tab. 1 Comparison between NCL method and three enzymatic strategies
| 项目 | SrtA转肽酶 | Butelase 1转肽酶 | Subtilisin人工连接酶 | NCL | |
|---|---|---|---|---|---|
| 最初来源物种 | 金黄色葡萄球菌Staphylococcus aureus | 蝶豆 Clitoria ternatea | 解淀粉芽孢杆菌 Bacillus amyloliquefaciens | —— | |
| 酶的获取与产量[ | SrtA转肽酶可通过商业渠道获得或大肠杆菌重组表达 >40 mg/L | Butelase 1转肽酶通过植物材料提取 约5 mg/kg | Omniligase-1可通过商业渠道获得或枯草芽孢杆菌重组表达 >500 mg/L | —— | |
| 底物活化 | 底物无需活化 | 底物无需活化 | 多肽C端活化为 氧酯或硫酯 | 多肽C端活化为 硫酯 | |
连接 位点 序列 | C端 | LPXT-G | N/D-HV | X4X3X2X1—,X2外均避免Pro,X4偏好疏水&芳香族氨基酸 | 无限制 |
| N端 | (G) n | —X1X2,其中X1避免Pro和 酸性氨基酸,X2一般为ILVC | —X1’X2’均避免Pro | Cys | |
| 连接“疤痕” | LPXT(G) n | 一个Asn/Asp残基 | 无痕(X4X3X2X1 X1’X2’) | 一个Cys残基 | |
应用 范围 | 多肽首 尾环化 | 短肽(大于19个氨基酸残基)和蛋白质皆可环化 | 短肽(大于9个氨基酸残基)和蛋白质皆可环化 | 环化多肽一般不超过40个残基 | 环化多肽一般不超过40个残基 |
蛋白末 端修饰 | 蛋白质N端与C端皆可 | 蛋白质N端与C端皆可 | 大多用于蛋白质N端修饰鲜有C端修饰 | 蛋白质N端与C端皆可 | |
蛋白质 全合成 | 尚无应用报道 | 尚无应用报道 | 可以应用 | 广泛应用 | |
| 1 | LAU J L, DUNN M K. Therapeutic peptides: historical perspectives, current development trends, and future directions [J]. Bioorganic & Medicinal Chemistry, 2018, 26(10): 2700-2707. |
| 2 | XIAO Y, JIE M, LI B, et al. Peptide-based treatment: a promising cancer therapy [J]. Journal of Immunology Research, 2015,2015: 761820. |
| 3 | VADEVOO S M P, GURUNG S, KHAN F, et al. Peptide-based targeted therapeutics and apoptosis imaging probes for cancer therapy [J]. Archives of Pharmacal Research, 2019, 42(2): 150-158. |
| 4 | TOPLAK A, NUIJENS T, QUAEDFLIEG P J L M, et al. Peptiligase, an enzyme for efficient chemoenzymatic peptide synthesis and cyclization in water [J]. Advanced Synthesis & Catalysis, 2016, 358: 2140-2147. |
| 5 | ZORZI A, DEYLE K, HEINIS C. Cyclic peptide therapeutics: past, present and future [J]. Current Opinion in Chemical Biology, 2017, 38: 24-29. |
| 6 | CHOW H Y, ZHANG Y, MATHESON E, et al. Ligation technologies for the synthesis of cyclic peptides [J]. Chemical Reviews, 2019, 119(17): 9971-10001. |
| 7 | WEEKS A M, WELLS J A. Subtiligase-catalyzed peptide ligation [J]. Chemical Reviews, 2020, 120(6): 3127-3160. |
| 8 | HOUEN G. Peptide antibodies[M]. New York: Humana Press, 2015: 33-50. |
| 9 | BEHRENDT R, WHITE P, OFFER J. Advances in Fmoc solid-phase peptide synthesis [J]. Journal of Peptide Science, 2016, 22(1): 4-27. |
| 10 | SCHMIDT M, TOPLAK A, QUAEDFLIEG P J L M, et al. Enzyme-mediated ligation technologies for peptides and proteins [J]. Current Opinion in Chemical Biology, 2017, 38: 1-7. |
| 11 | ZHANG Y, PARK K, SUAZO K F, et al. Recent progress in enzymatic protein labelling techniques and their applications [J]. Chemical Society Reviews, 2018, 47(24): 9106-9136. |
| 12 | AGOURIDAS V, MAHDI O E, DIEMER V, et al. Native chemical ligation and extended methods: mechanisms, catalysis, scope, and limitations [J]. Chemical Reviews, 2019, 119(12): 7328-7443. |
| 13 | CONIBEAR A C, WATSON E E, PAYNE R J, et al. Native chemical ligation in protein synthesis and semi-synthesis [J]. Chemical Society Reviews, 2018, 47(24):9046-9068. |
| 14 | TAM A, SOELLNER M B, RAINES R T. Water-soluble phosphinothiols for traceless Staudinger ligation and integration with expressed protein ligation [J]. Journal of the American Chemical Society, 2007, 129(37): 11421-11430. |
| 15 | PUSTERLA I, BODE J W. The mechanism of the a-ketoacid-hydroxylamine amide-forming ligation [J]. Angewandte Chemie International Edition, 2012, 51: 513-516. |
| 16 | ZHANG Y F, XU C, LAM H Y, et al. Protein chemical synthesis by serine and threonine ligation [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(17): 6657-6662. |
| 17 | KULKARNI S S, WATSON E E, PREMDJEE B, et al. Diselenide-selenoester ligation for chemical protein synthesis [J]. Nature Protocols, 2019, 14: 2229-2257. |
| 18 | DAWSON P E, MUIR T W, CLARK-LEWIS I, et al. Synthesis of proteins by native chemical ligation [J]. Science, 1994, 266(5186):776-779. |
| 19 | ZHENG J, TANG S, QI Y, et al. Chemical synthesis of proteins using peptide hydrazides as thioester surrogates [J]. Nature Protocols, 2013, 8: 2483-2495. |
| 20 | FLOOD D T, HINTZEN J C J, BIRD M J, et al. Leveraging the Knorr pyrazole synthesis for the facile generation of thioester surrogates for use in native chemical ligation [J]. Angewandte Chemie International Edition, 2018, 57: 11634-11639. |
| 21 | MUIR T W, SONDHI D, COLE P A. Expressed protein ligation: a general method for protein engineering [J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(12): 6705-6710. |
| 22 | MALINS L R, PAYNE R J. Recent extensions to native chemical ligation for the chemical synthesis of peptides and proteins [J]. Current Opinion in Chemical Biology, 2014, 22: 70-78. |
| 23 | VILA-PERELLÓ M, LIU Z, SHAH N H, et al. Streamlined expressed protein ligation using split inteins [J]. Journal of the American Chemical Society, 2013, 135: 286-292. |
| 24 | MAZMANIAN S K, LIU G, TON-THAT H, et al. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall [J]. Science, 1999, 285(5428): 760-763. |
| 25 | ATONS J M, CHEW G L, GUIMARAES C P, et al. Site-specific N-and C-terminal labeling of a single polypeptide using sortases of different specificity [J]. Journal of the American Chemical Society, 2009, 131(31): 10800-10801. |
| 26 | NUIJENS T, TOPLAK A, SCHMIDT M, et al. Natural occurring and engineered enzymes for peptide ligation and cyclization [J]. Frontiers in Chemistry, 2019, 7: 829. |
| 27 | MAO H, HART S A, SCHINK A, et al. Sortase-mediated protein ligation: a new method for protein engineering [J]. Journal of the American Chemical Society, 2004, 126(9): 2670-2671. |
| 28 | TANAKA T, YAMAMOTO T, TSUKIJI S, et al. Site-specific protein modification on living cells catalyzed by sortase [J]. ChemBioChem, 2008, 9: 802-807. |
| 29 | YAMAMOTO T, NAGAMUNE T. Expansion of the sortase-mediated labeling method for site-specific N-terminal labeling of cell surface proteins on living cells [J]. Chemical Communications, 2009, 9: 1022-1024. |
| 30 | HESS G T, CRAGNOLINI J J, POPP M W, et al. M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins [J]. Bioconjugate Chemistry, 2012, 23(7): 1478-1487. |
| 31 | VAN'T HOF W, MAŇÁSKOVÁ S H, VEERMAN E C I, et al. Sortase-mediated backbone cyclization of proteins and peptides [J]. Biological Chemistry, 2015, 396(4): 283-293. |
| 32 | BOLSCHER J G M, OUDHOFF M J, NAZMI K, et al. Sortase A as a tool for high-yield histatin cyclization [J]. The FASEB Journal, 2011, 25(8): 2650-2658. |
| 33 | STANGER K, MAURER T, KALUARACHCHI H, et al. Backbone cyclization of a recombinant cystine-knot peptide by engineered Sortase A [J]. FEBS Letters, 2014, 588(23): 4487-4496. |
| 34 | ZHANG J, YAMAGUCHI S, NAGAMUNE T. Sortase A-mediated synthesis of ligand-grafted cyclized peptides for modulating a model protein-protein interaction [J]. Biotechnology Journal, 2015, 10: 1499-1505. |
| 35 | SCHMOHL L, SCHWARZER D. Sortase-mediated ligations for the site-specific modification of proteins [J]. Current Opinion in Chemical Biology, 2014, 22: 122-128. |
| 36 | SCHMIDT M, TOPLAK A, QUAEDFLIEG P J L M, et al. Enzyme-catalyzed peptide cyclization [J]. Drug Discovery Today. Technologies, 2017, 26: 11-16. |
| 37 | THOMPSON R E, STEVENS A J, MUIR T W. Protein engineering through tandem transamidation [J]. Nature Chemistry, 2019, 11: 737-743. |
| 38 | LI Y, LI Y, PAN M, et al. Irreversible site-specific hydrazinolysis of proteins by use of sortase [J]. Angewandte Chemie International Edition, 2014, 53: 2198-2202. |
| 39 | NGUYEN G K T, WANG S J, QIU Y B, et al. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis [J]. Nature Chemical Biology, 2014, 10(9): 732-738. |
| 40 | NGUYEN G K T, KAM A, LOO S, et al. Butelase 1: a versatile ligase for peptide and protein macrocyclization [J]. Journal of the American Chemical Society, 2015, 137(49): 15398-15401. |
| 41 | NGUYEN G K T, QIU Y B, CAO Y, et al. Butelase-mediated cyclization and ligation of peptides and proteins [J]. Nature Protocols, 2016, 11(10): 1977-1988. |
| 42 | HEMU X, QIU Y B, NGUYEN G K T, et al. Total synthesis of circular bacteriocins by Butelase 1 [J]. Journal of the American Chemical Society, 2016, 138(22): 6968-6971. |
| 43 | HEMU X, ZHANG X H, TAM J P. Ligase-controlled cyclo-oligomerization of peptides [J]. Organic Letters, 2019, 21(7): 2029-2032. |
| 44 | NGUYEN G K T, CAO Y, WANG W, et al. Site-specific N-terminal labeling of peptides and proteins using butelase 1 and thiodepsipeptide [J]. Angewandte Chemie International Edition, 2015, 54: 15694-15698. |
| 45 | BI X, YIN J, NGUYEN G K T, et al. Enzymatic engineering of live bacterial cell surfaces using butelase 1 [J]. Angewandte Chemie International Edition, 2017, 56: 7822-7825. |
| 46 | CAO Y, NGUYEN G K T, CHUAH S, et al. Butelase-mediated ligation as an efficient bioconjugation method for the synthesis of peptide dendrimers [J]. Bioconjugate Chemistry, 2016, 27(11): 2592-2596. |
| 47 | CAO Y, NGUYEN G K T, TAM J P, et al. Butelase-mediated synthesis of protein thioesters and its application for tandem chemoenzymatic ligation [J]. Chemical Communications, 2015, 51(97): 17289-17292. |
| 48 | NUIJENS T, SCHMIDT M. Butelase 1-mediated ligation of peptides and proteins [M]. New York: Humana Press, 2019: 83-109. |
| 49 | HARRIS K S, DUREK T, KAAS Q, et al. Efficient backbone cyclization of linear peptides by a recombinant asparaginyl endopeptidase [J]. Nature Communications, 2015, 6: 10199. |
| 50 | YANG R, WONG Y H, NGUYEN G K T, et al. Engineering a catalytically efficient recombinant protein ligase [J]. Journal of the American Chemical Society, 2017, 139: 5351-5358. |
| 51 | POLGARF L, BENDER M L. The reactivity of thiol-subtilisin, an enzyme containing a synthetic functional group [J]. Biochemistry, 1967, 6(2): 610-620. |
| 52 | NAKATSUKA T, SASAKI T, KAISER E T. Peptide segment coupling catalyzed by the semisynthetic enzyme thiolsubtilisin [J]. Journal of the American Chemical Society, 1987, 109(12): 3808-3810. |
| 53 | ABRAHMSÉN L, TOM J, BURNIER J, et al. Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution [J]. Biochemistry, 1991, 30(17): 4151-4159. |
| 54 | CHANG T K, JACKSON D Y, BURNIER J P, et al. Subtiligase: a tool for semisynthesis of proteins [J]. Proceedings of the National Academy of Sciences of the United States of America, 1994, 91(26): 12544-12548. |
| 55 | ATWELL S, WELLS J A. Selection for improved subtiligases by phage display [J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(17): 9497-9502. |
| 56 | WEEKS A M, WELLS J A. Engineering peptide ligase specificity by proteomic identification of ligation sites [J]. Nature Chemical Biology, 2018, 14(1): 50-57. |
| 57 | JACKSON D Y, BURNIER J, QUAN C, et al. A designed peptide ligase for total synthesis of ribonuclease a with unnatural catalytic residues [J]. Science, 1994, 266(5183): 243-247. |
| 58 | JACKSON D Y, BUMIER J P, WELLS J A. Enzymatic cyclization of linear peptide esters using subtiligase [J]. Journal of the American Chemical Society, 1995, 117: 819-820. |
| 59 | HENAGER S H, CHU N, CHEN Z, et al. Enzyme-catalyzed expressed protein ligation[J]. Nature Methods, 2016, 13(11): 925-927. |
| 60 | MAHRUS S, TRINIDAD J C, BARKAN D T, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini [J]. Cell, 2008, 134: 866-876. |
| 61 | NUIJENS T, TOPLAK A, QUAEDFLIEG P J L M, et al. Engineering a diverse ligase toolbox for peptide segment condensation [J]. Advanced Synthesis & Catalysis, 2016, 358(24): 4041-4048. |
| 62 | SCHMIDT M, TOPLAK A, QUAEDFLIEG P J L M, et al. Omniligase-1: a powerful tool for peptide head-to-tail cyclization [J]. Advanced Synthesis & Catalysis, 2017, 359: 2050-2055. |
| 63 | PAWLAS J, NUIJENS T, PERSSON B, et al. Sustainable, cost-efficient manufacturing of therapeutic peptides using chemo-enzymatic peptide synthesis (CEPS) [J]. Green Chemistry, 2019, 21: 6451-6467. |
| 64 | SCHMIDT M, HUANG Y, OLIVEIRA E F T, et al. Efficient enzymatic cyclization of disulfide-rich peptides using peptiligases [J]. ChemBioChem, 2019, 20(12): 1524-1529. |
| 65 | RICHELLE G J J, SCHMIDT M, IPPEL H, et al. A one-pot "triple-c'' multicyclization methodology for the synthesis of highly constrained isomerically pure tetracyclic peptides [J]. ChemBioChem, 2018, 19(18): 1934-1938. |
| 66 | STREEFKERK D E, SCHMIDT M, IPPEL J H, et al. Synthesis of constrained tetracyclic peptides by consecutive CEPS, CLIPS, and oxime ligation [J]. Organic Letters, 2019, 21: 2095-2100. |
| 67 | SCHMIDT M, TOPLAK A, ROZEBOOM H J, et al. Design of a substrate-tailored peptiligase variant for the efficient synthesis of thymosin-α1 [J]. Organic & Biomolecular Chemistry, 2018, 16: 609-618. |
| 68 | HENNINOT A, COLLINS J C, NUSS J M. The current state of peptide drug discovery: back to the future? [J]. Journal of Medicinal Chemistry, 2018, 61: 1382-1414. |
| 69 | HUANG P, BOYKEN S E, BAKER D. The coming of age of de novo protein design [J]. Nature, 2016, 537: 320-327. |
| 70 | MILLS J H, KHARE S D, BOLDUC J M, et al. Computational design of an unnatural amino acid dependent metalloprotein with atomic level accuracy [J]. Journal of the American Chemical Society, 2013, 135(36): 13393-13399. |
| [1] | LIU Wanqiu, JI Xiangyang, XU Huiling, LU Yicong, LI Jian. Cell-free protein synthesis system enables rapid and efficient biosynthesis of restriction endonucleases [J]. Synthetic Biology Journal, 2023, 4(4): 840-851. |
| [2] | TANG Shiming, HU Jiyuan, ZHENG Suiping, HAN Shuangyan, LIN Ying. Designing, building and rapid prototyping of biosynthesis module based on cell-free system [J]. Synthetic Biology Journal, 2022, 3(6): 1250-1261. |
| [3] | HOU Jiaqi, JIANG Nan, MA Lianju, LU Yuan. Cell-free protein synthesis: from basic research to engineering applications [J]. Synthetic Biology Journal, 2022, 3(3): 465-486. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||