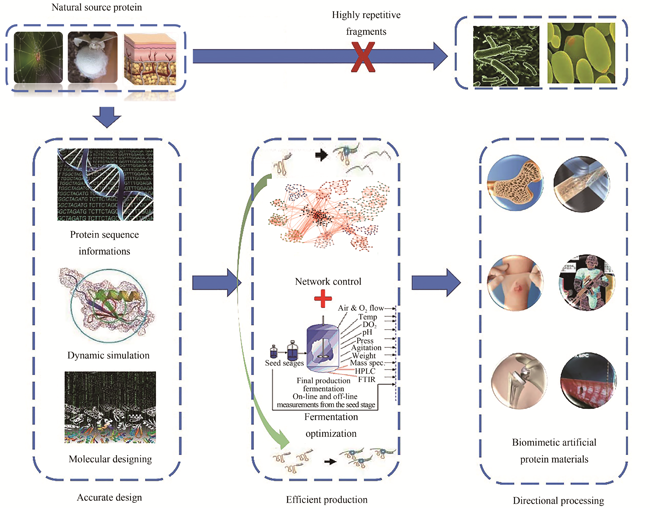
Protein functional materials have good biocompatibility, degradability and versatility, so they have great application values in the fields of medicine, military and textile. However, due to their unique characteristics of high molecular weight, high-frequency amino acids and special post-translation modifications, there are bottlenecks in its low expression rate in artificial cell synthesis, poor compatibility between functional elements and chassis cells, and unstable structure and efficacy, which seriously limit the efficient production and application of these proteins. Synthetic biology, as the third biotechnology revolution, has the advantages of renewable resources, low pollution, easy control and directional design of biological macromolecules. With the gradual excavation and analysis of the functional principles and novel design concepts of protein functional materials, and the development of alternative materials with excellent performance under specific conditions, it has brought revolutionary changes to human social life. It has been reported that such protein functional materials have great application value in cancer diagnosis and treatment, regenerative medicine, gene delivery system, data storage and so on. Although synthetic biology has broadened the range of potential applications of protein functional materials, there are still some limitations. This requires us to carry out research from two different perspectives, biology and materials science. On the one hand, we should establish a common technology platform to form a complete upper, middle and downstream research system. On the other hand, we should establish targeted research based on the characteristics of different protein materials to produce materials with better performance. This article introduces the application and development of protein functional materials. Taking the efficient protein synthesis and functional requirements as the guide, the synthesis strategy of protein functional materials are explained from the aspects of protein molecule orientation design, cell factory construction and adaptation control, and protein material processing applications, in order to realize their functional directional enhancement and industrial production. The limitations of synthetic biology oriented protein functional materials in biology and materials science are prospected, which lay a foundation for the wide application of protein functional materials with excellent properties.