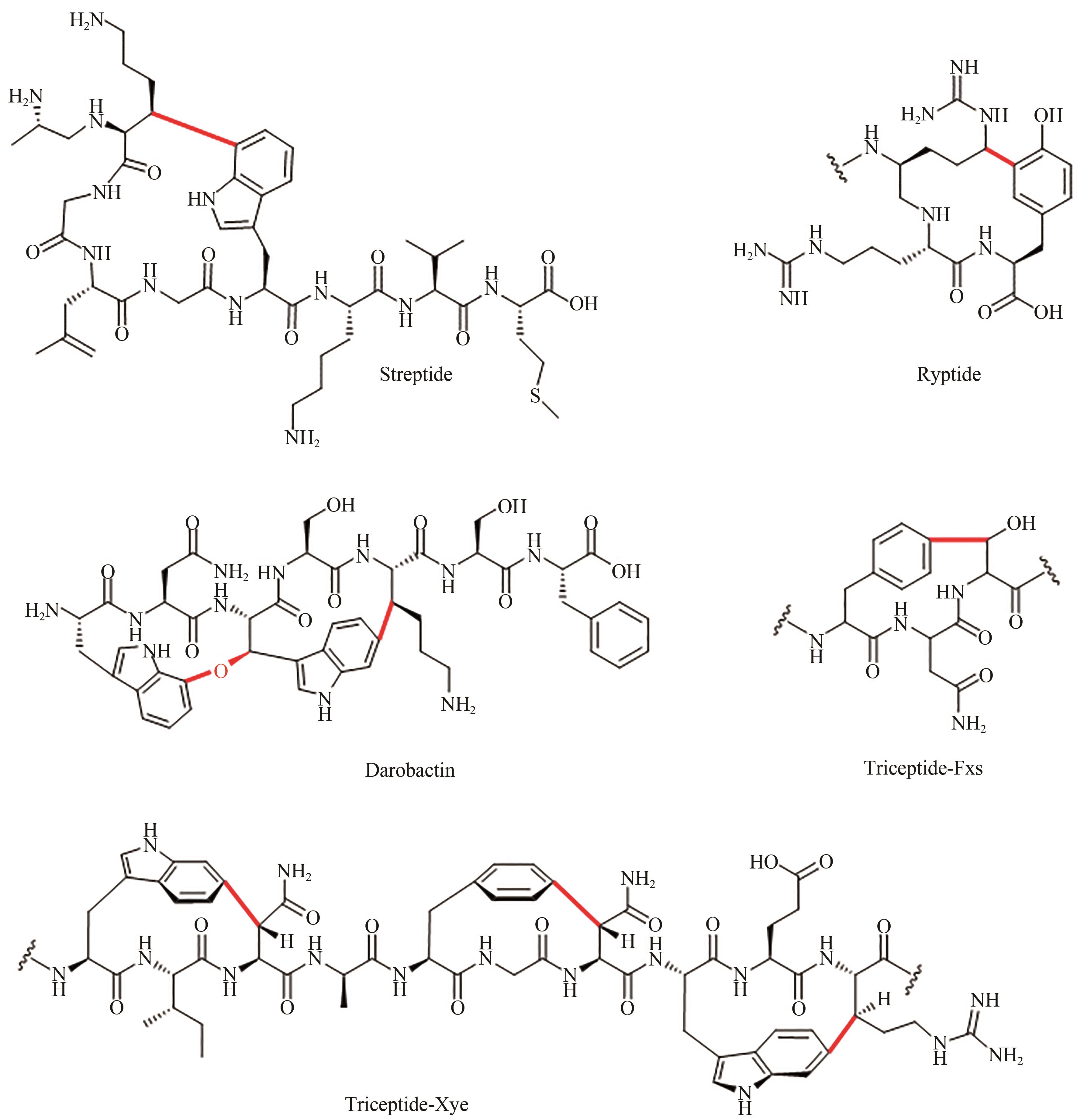
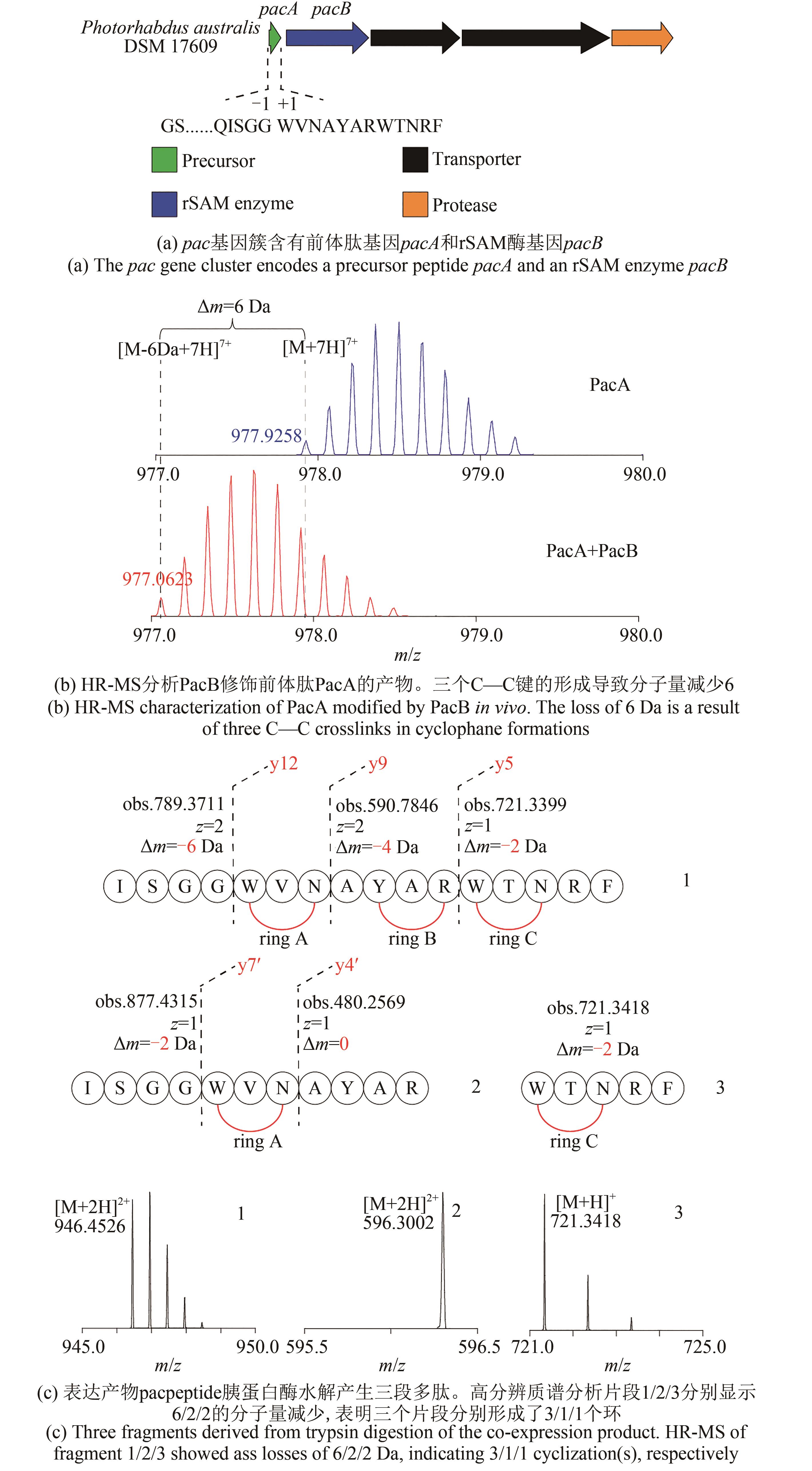
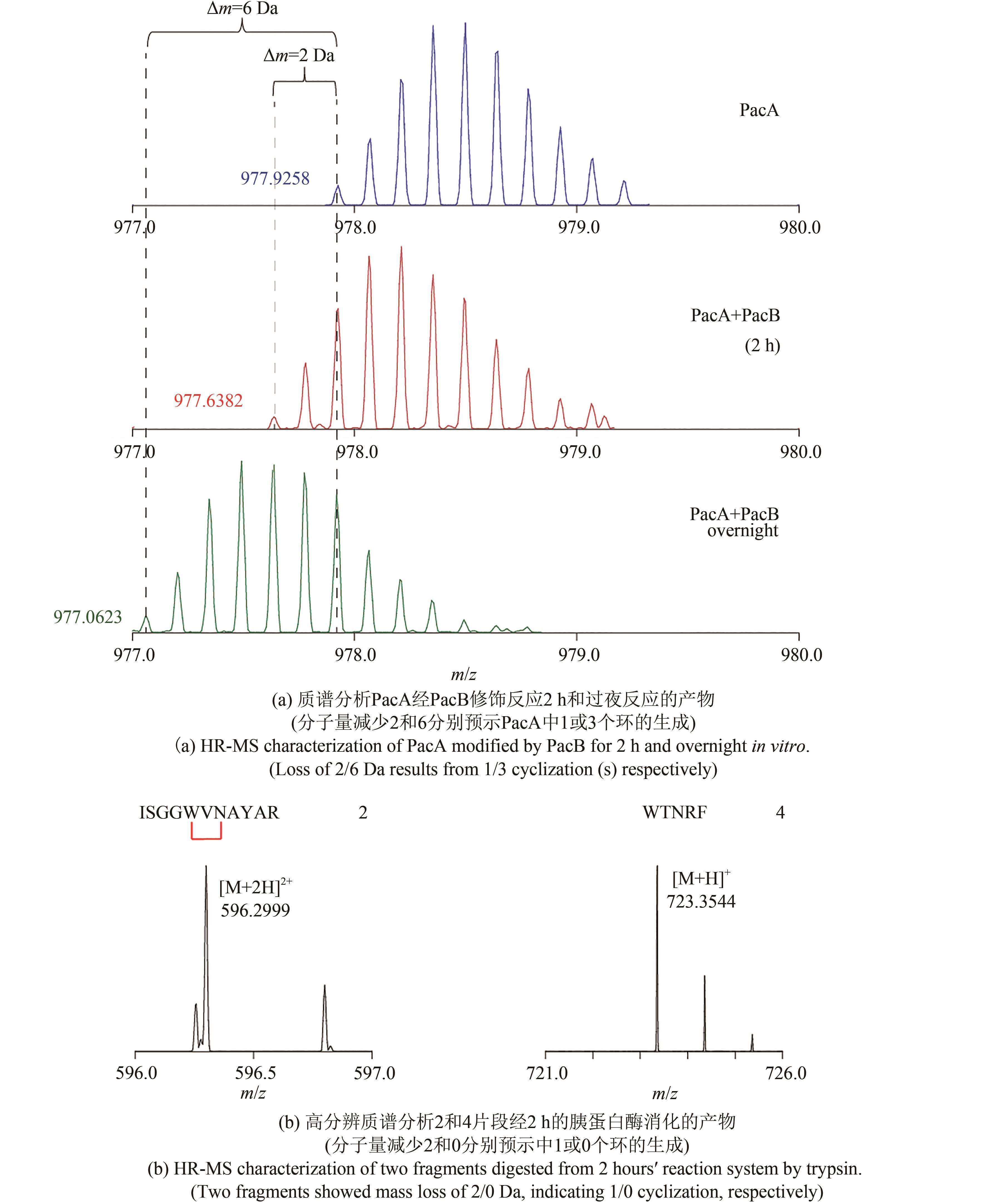
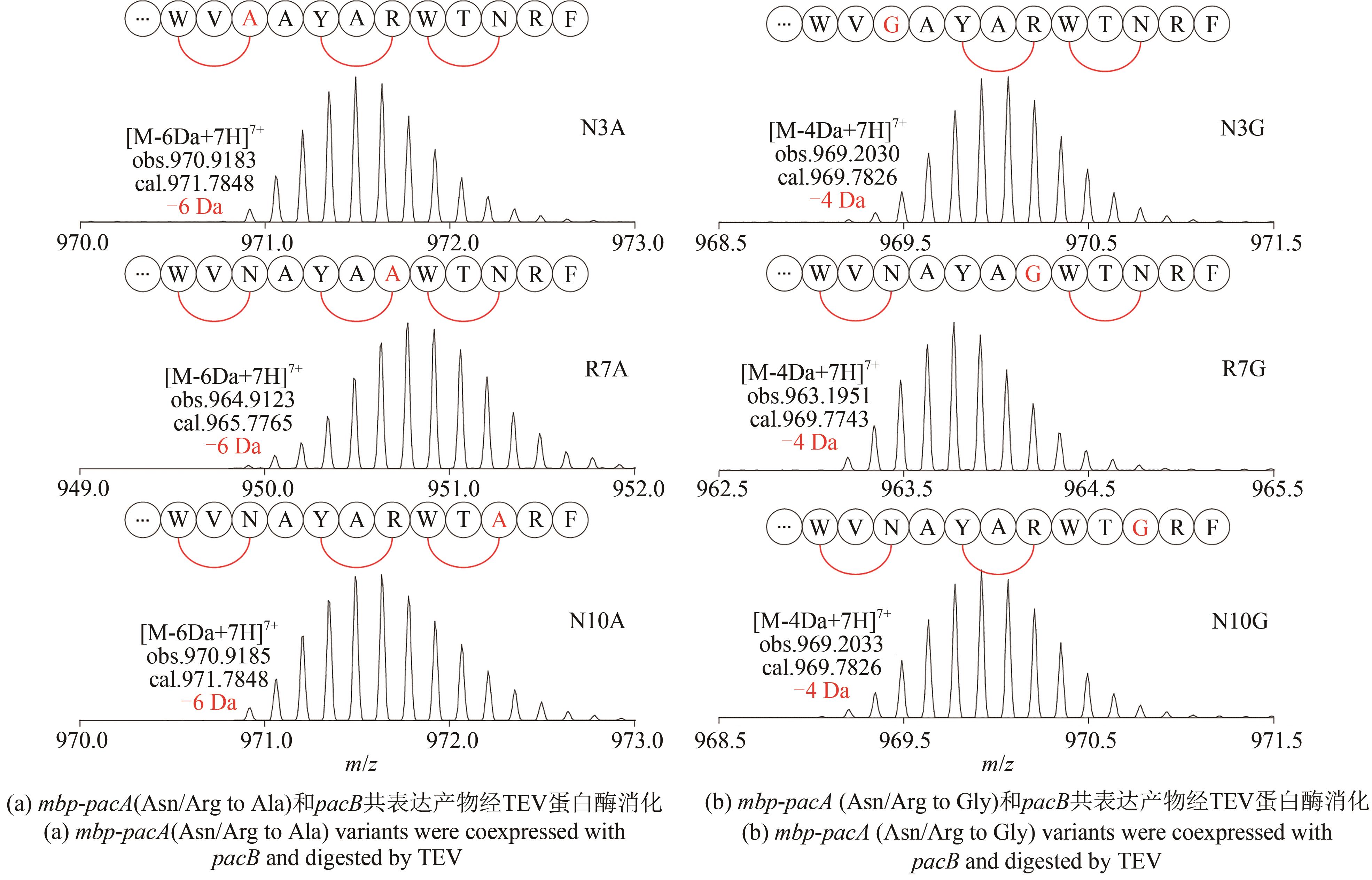
| 1 |
ARNISON P G, BIBB M J, BIERBAUM G, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature[J]. Natural Product Reports, 2013, 30(1): 108-160.
|
| 2 |
MONTALBÁN-LÓPEZ M, SCOTT T A, RAMESH S, et al. New developments in RiPP discovery, enzymology and engineering[J]. Natural Product Reports, 2021, 38(1): 130-239.
|
| 3 |
LU J X, LI Y Q, BAI Z B, et al. Enzymatic macrocyclization of ribosomally synthesized and posttranslational modified peptides via C-S and C-C bond formation[J]. Natural Product Reports, 2021, 38(5): 981-992.
|
| 4 |
DONK W A VAN DER, NAIR S K. Structure and mechanism of lanthipeptide biosynthetic enzymes[J]. Current Opinion in Structural Biology, 2014, 29: 58-66.
|
| 5 |
ZHANG Q, YU Y, VÉLASQUEZ J E, et al. Evolution of lanthipeptide synthetases[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(45): 18361-18366.
|
| 6 |
KRAAIJ C VAN, DE VOS W M, SIEZEN R J, et al. Lantibiotics: biosynthesis, mode of action and applications[J]. Natural Product Reports, 1999, 16(5): 575-587.
|
| 7 |
FLUHE L, MARAHIEL M A. Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis[J]. Current Opinion in Structural Biology, 2013, 17(4): 605-612.
|
| 8 |
CHEN Y L, WANG J X, LI G Q, et al. Current advancements in sactipeptide natural products[J]. Frontiers in Chemistry, 2021, 9: 595991.
|
| 9 |
MO T L, JI X J, YUAN W, et al. Thuricin Z: A narrow-spectrum sactibiotic that targets the cell membrane[J]. Angewandte Chemie International Edition, 2019, 58(52): 18793-18797.
|
| 10 |
DALY N L, CRAIK D J. Bioactive cystine knot proteins[J]. Current Opinion in Structural Biology, 2011, 15: 362-368.
|
| 11 |
CHEKAN J R, ESTRADA P, COVELLO P S, et al. Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(25): 6551-6556.
|
| 12 |
BURNETT P G, JADHAV P D, OKINYO-OWITI D P, et al.Glycine-containing flaxseed orbitides[J]. Journal of Natural Products, 2015, 78: 681-688.
|
| 13 |
SHIM Y Y, YOUNG L W, ARNISON P G, et al. Proposed systematic nomenclature for orbitides[J]. Journal of Natural Products, 2015, 78(4): 645-652.
|
| 14 |
DE VEER S J, KAN M W, CRAIK D J. Cyclotides: from structure to function[J]. Chemical Reviews, 2019, 119(24): 12375-12421.
|
| 15 |
BURMAN R, GUNASEKERA S, STRÖMSTEDT A A, et al. Chemistry and biology of cyclotides: circular plant peptides outside the box[J]. Journal of Natural Products, 2014, 77(3): 724-736.
|
| 16 |
CRAIK D J, CEMAZAR M, DALY N L. The chemistry and biology of cyclotides[J]. Current Opinion in Drug Discovery & Development, 2007, 10(2): 176-184.
|
| 17 |
MARTINS J, VASCONCELOS V. Cyanobactins from cyanobacteria: current genetic and chemical state of knowledge[J]. Marine Drugs, 2015, 13(11): 6910-6946.
|
| 18 |
HUDSON G A, MITCHELL D A. RiPP antibiotics: biosynthesis and engineering potential[J]. Current Opinion in Microbiology, 2018, 45: 61-69.[PubMed]
|
| 19 |
DONIA M S, RAVEL J, SCHMIDT E W. A global assembly line for cyanobactins[J]. Nature Chemical Biology, 2008, 4(6): 341-343.
|
| 20 |
VINOGRADOV A A, SUGA H. Introduction to thiopeptides: biological activity, biosynthesis, and strategies for functional reprogramming[J]. Cell Chemical Biology, 2020, 27(8): 1032-1051.
|
| 21 |
MA S, CHEN H, LI H, et al. Post-translational formation of aminomalonate by a promiscuous peptide-modifying radical SAM enzyme[J]. Angewandte Chemie International Edition, 2021, 60(36): 19957-19964.
|
| 22 |
NGUYEN T Q N, TOOH Y W, SUGIYAMA R, et al.Post-translational formation of strained cyclophanes in bacteria[J]. Nature Chemistry, 2020, 12: 1042-1053.
|
| 23 |
SCHRAMMA K R, BUSHIN L B, SEYEDSAYAMDOST M R. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink[J]. Nature Chemistry, 2015, 7(5): 431-437.
|
| 24 |
KAUR H, JAKOB R P, MARZINEK J K, et al.The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase[J]. Nature, 2021, 593: 125-129.
|
| 25 |
BUSHIN L B, SEYEDSAYAMDOST M R. Guidelines for determining the structures of radical SAM enzyme catalyzed modifications in the biosynthesis of RiPP natural products[J]. Methods in Enzymology, 2018, 606: 439-460.
|
| 26 |
BENJDIA A, BALTY C, BERTEAU O. Radical SAM enzymes in the biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPPs)[J]. Frontiers in Chemistry, 2017, 5: 87.
|
| 27 |
ZHANG Q, ORTEGA M, SHI Y X, et al. Structural investigation of ribosomally synthesized natural products by hypothetical structure enumeration and evaluation using tandem MS[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(33): 12031-12036.
|
| 28 |
STROHALM M, HASSMAN M, KOSATA B, et al. mMass data miner: an open source alternative for mass spectrometric data analysis[J]. Rapid Communications in Mass Spectrometry, 2008, 22(6): 905-908.
|