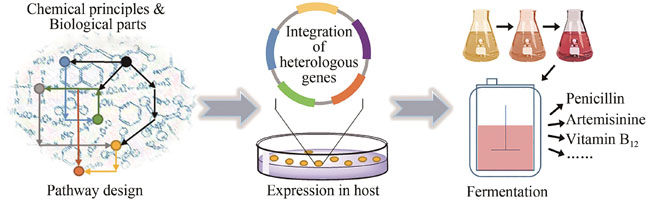
The complexity and diversity of natural products have made them a rich source for drug and agrochemical discovery. To overcome the supplying limitation of natural resources, tremendous effort has been made by the academic and industrial communities during the past two centuries for the total artificial synthesis of natural products. In this regard, total chemical synthesis has achieved significant progress, and numerous highly complex natural products have been synthesized through different chemical processes. Despite these great achievements in total chemical synthesis, there are still many challenges including expensive chemical reagents, harsh reaction conditions, difficult control on stereoselectivity, long synthetic route, and low product yield. Notably, the development of synthetic biology has allowed more and more natural products to be produced through biological cell factories, which provides a new and complementary strategy for the synthesis of natural products at a large scale. This review critically comments on the representative advances in total chemical synthesis of natural products (Section 1), and then highlight major progress and trends in the biosynthesis of pharmaceutically important natural products (Sections 2 and 3). In Section 2.1, we selected the production of penicillin, erythromycin, and avermectin as examples to analyze the modification and optimization of natural product biosynthetic pathways. The discovery and utilization of secondary metabolites from microorganisms has been a continuous driving force in the field of natural products. Notably, significant progress has been made in the total biosynthesis of natural products from secondary metabolism via the genetic manipulation of microbial cells. In Section 2.2, we selected Vitamin B12 and Tropane alkaloids as examples to demonstrate the use of heterologous expression and biological production for natural product synthesis. In recent years, on the basis of analyzing the structure of natural products in animals, plants, and microorganisms, great advances have emerged in exploring their biochemical reaction mechanisms and synthetic routes. More importantly, expressing and regulating the relative genes in heterologous microbial cells have enabled the complete biosynthesis of many natural products. Furthermore, in Section 3, human insulin, artemisinin, saframycin, azaphilone, kainic acid, and podophyllotoxin were selected as examples to showcase the power of merging chemical and biological processes for the total synthesis of natural products. Although there are still many challenges in the total synthesis of new and complex natural products, biosynthesis will ultimately play a significant role in the construction of natural molecules and their relative analogues. By taking advantage of the merits with organic chemistry, synthetic biology, and artificial intelligence, the development of highly efficient and automatic biosynthesis could be a trend in this field.