Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (5): 804-814.DOI: 10.12211/2096-8280.2021-081
Previous Articles Next Articles
Regulation on oxidation selectivity for β-amyrin by Class Ⅱ cytochrome P450 enzymes
SUN Wentao1, ZHANG Xinzhe2, WAN Shengtong2, WANG Ruwen2, LI Chun1,2
- 1.Department of Chemical Engineering,Tsinghua University,Beijing 100084,China
2.School of Chemistry and Chemical Engineering,Beijing Institute of Technology,Beijing 100101,China
-
Received:2021-08-06Revised:2021-09-24Online:2021-11-19Published:2021-10-31 -
Contact:LI Chun
Ⅱ型细胞色素P450酶氧化β-香树脂醇的选择性调控研究
孙文涛1, 张昕哲2, 万盛通2, 王茹雯2, 李春1,2
- 1.清华大学化工系,北京 100084
2.北京理工大学化学与化工学院,北京 100101
-
通讯作者:李春 -
作者简介:孙文涛 (1988—),男,博士,博士后。研究方向为酶工程与合成生物学。E-mail:sunwentao@tsinghua.edu.cn李春 (1970—),男,博士生导师,教授。研究方向为生物催化与酶工程,合成生物学与代谢工程。E-mail:lichun@tsinghua.edu.cn -
基金资助:国家重点研发计划(2018YFA0901800);国家自然科学基金(21736002);清华大学“水木学者”计划(2020SM097)
CLC Number:
Cite this article
SUN Wentao, ZHANG Xinzhe, WAN Shengtong, WANG Ruwen, LI Chun. Regulation on oxidation selectivity for β-amyrin by Class Ⅱ cytochrome P450 enzymes[J]. Synthetic Biology Journal, 2021, 2(5): 804-814.
孙文涛, 张昕哲, 万盛通, 王茹雯, 李春. Ⅱ型细胞色素P450酶氧化β-香树脂醇的选择性调控研究[J]. 合成生物学, 2021, 2(5): 804-814.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-081
| 菌株 | 基因型 | 来源 |
|---|---|---|
| SynV | MATa his3Δ1;leu2;met15Δ;ura3-52 | 天津大学 |
| SynV8 | SynV: ΔGal80 | 本研究构建 |
| GA0-1 | SynV8: ΔHO:: FBA1p-bAS-CYC1-ENO2p-Uni25647-TYS1t-PGK1p-GuCPR1-PGK1t-G418 | 本研究构建 |
| GA0 | SynV8: ΔHO:: FBA1p-bAS-CYC1t-Gal2p-CYP72A63-ADH1t-ENO2p-Uni25647-TYS1t-PGK1p-GuCPR1-PGK1t-HIS3 | 本研究构建 |
| GA160 | SynV8: ΔHO:: FBA1p-bAS-CYC1t-Gal2p-CYP72A63(T338S)-ADH1t-ENO2p-Uni25647-TYS1t-PGK1p-GuCPR1-PGK1t-HIS3 | 本研究构建 |
| GA180-1 | SynV8: ΔHO:: FBA1p-bAS-CYC1t-Gal2p-CYP72A63(T338S)-ADH1t-Gal7p-Uni25647-TYS1t-PGK1p-GuCPR1-PGK1t-HIS3 | 本研究构建 |
Tab. 1 Strains used in this study
| 菌株 | 基因型 | 来源 |
|---|---|---|
| SynV | MATa his3Δ1;leu2;met15Δ;ura3-52 | 天津大学 |
| SynV8 | SynV: ΔGal80 | 本研究构建 |
| GA0-1 | SynV8: ΔHO:: FBA1p-bAS-CYC1-ENO2p-Uni25647-TYS1t-PGK1p-GuCPR1-PGK1t-G418 | 本研究构建 |
| GA0 | SynV8: ΔHO:: FBA1p-bAS-CYC1t-Gal2p-CYP72A63-ADH1t-ENO2p-Uni25647-TYS1t-PGK1p-GuCPR1-PGK1t-HIS3 | 本研究构建 |
| GA160 | SynV8: ΔHO:: FBA1p-bAS-CYC1t-Gal2p-CYP72A63(T338S)-ADH1t-ENO2p-Uni25647-TYS1t-PGK1p-GuCPR1-PGK1t-HIS3 | 本研究构建 |
| GA180-1 | SynV8: ΔHO:: FBA1p-bAS-CYC1t-Gal2p-CYP72A63(T338S)-ADH1t-Gal7p-Uni25647-TYS1t-PGK1p-GuCPR1-PGK1t-HIS3 | 本研究构建 |
| 成分 | 体积/μL |
|---|---|
| 1 mol/L Tris-HCl(pH7.5) | 3000 |
| 2 mol/L MgCl2 | 150 |
| dNTP mix | 240 |
| 1 mol/L DTT | 300 |
| PEG-8000 | 1.5 g |
| 0.1 mol/L NAD | 300 |
| ddH2O | 定容至6 mL,-80 ℃分装,保存 |
Tab. 2 Components of the 5×ISO buffer
| 成分 | 体积/μL |
|---|---|
| 1 mol/L Tris-HCl(pH7.5) | 3000 |
| 2 mol/L MgCl2 | 150 |
| dNTP mix | 240 |
| 1 mol/L DTT | 300 |
| PEG-8000 | 1.5 g |
| 0.1 mol/L NAD | 300 |
| ddH2O | 定容至6 mL,-80 ℃分装,保存 |
| 成分 | 体积/μL |
|---|---|
| 5×ISO buffer | 320 |
| 10 U/μL T5 exonuclease | 0.64 |
| 2 U/μL Phusion polymerase | 20 |
| 40 U/μL | 160 |
| ddH2O | 1200 |
Tab. 3 Components of Gibson reaction solution
| 成分 | 体积/μL |
|---|---|
| 5×ISO buffer | 320 |
| 10 U/μL T5 exonuclease | 0.64 |
| 2 U/μL Phusion polymerase | 20 |
| 40 U/μL | 160 |
| ddH2O | 1200 |

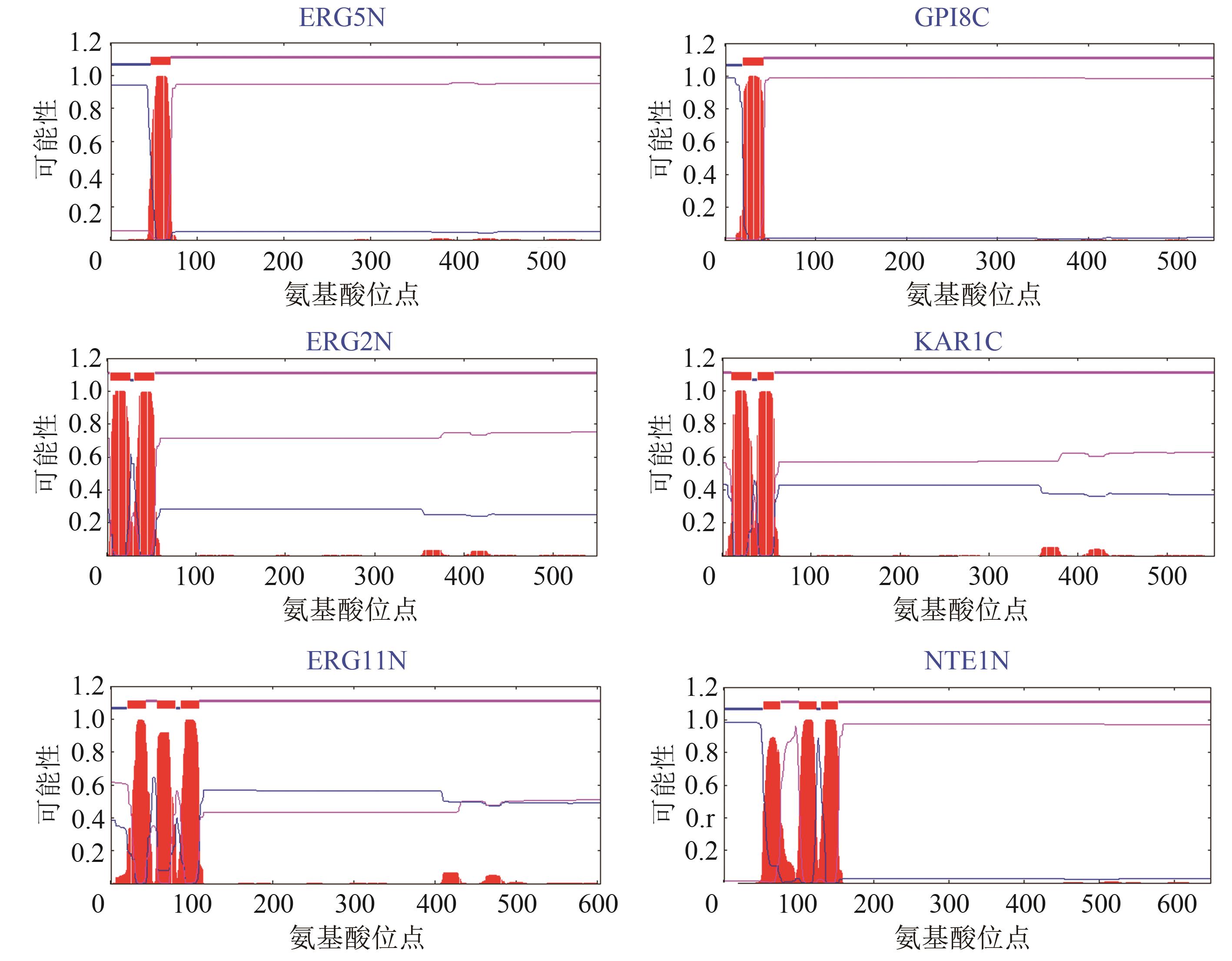
Fig. 2 Influence of the transmembrane structures on the substrate selectivity of CYP72A63 (T338S)[The same letters indicate no significant difference at the95% confidence level (P < 0.05)]
| 基因 | 注释 | FPKM Sgib | FPKM SynV |
|---|---|---|---|
| FAA1 | long-chain fatty acid-CoA ligase FAA1 | 1534.17 | 410.28 |
| SUR2 | sphingosine C-4 hydroxylase | 480.16 | 162.61 |
Tab. 4 Transcription levels of genes associated to lipid metabolism in different strains
| 基因 | 注释 | FPKM Sgib | FPKM SynV |
|---|---|---|---|
| FAA1 | long-chain fatty acid-CoA ligase FAA1 | 1534.17 | 410.28 |
| SUR2 | sphingosine C-4 hydroxylase | 480.16 | 162.61 |

Fig. 3 Product profile of the strains with FAA1or SUR2 knock out[The same letters indicate no significant difference at the 95% confidence level (P < 0.05)]
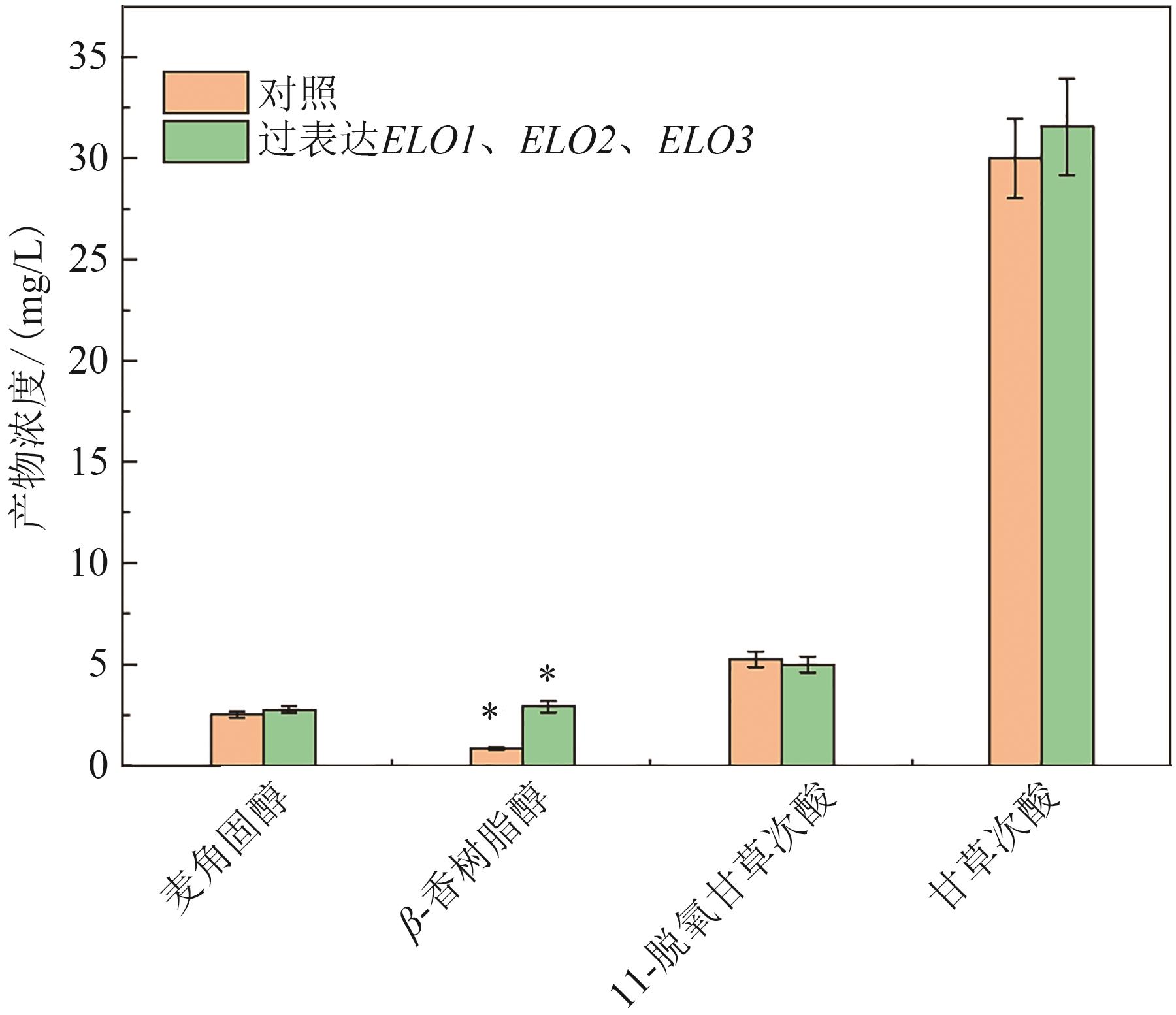
Fig. 4 Production of triterpenoids in the engineered strains overexpressing ELO1, ELO2 and ELO3[Asterisk (*) indicates significant difference at the 95% confidence level (P < 0.05)]
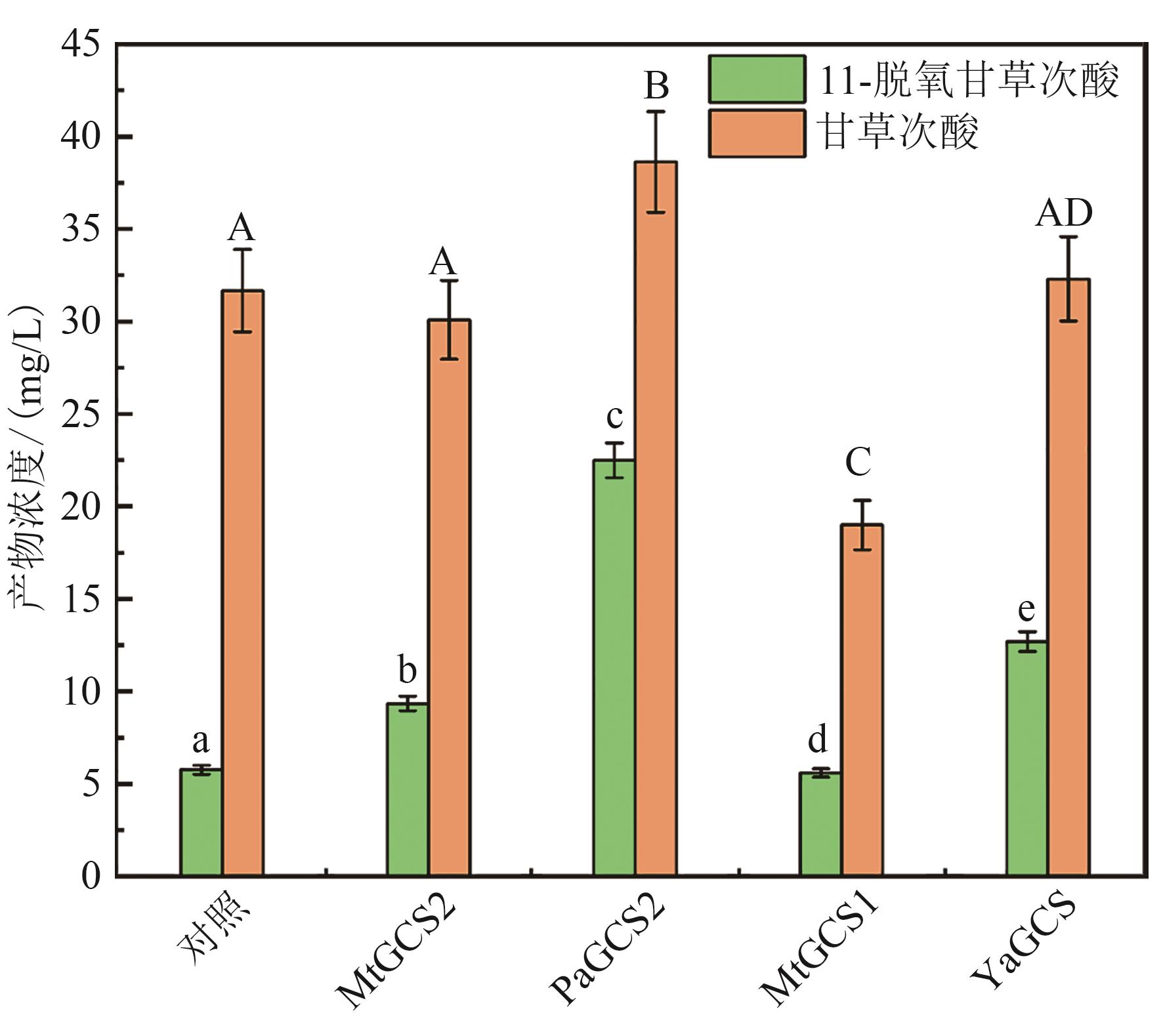
Fig. 5 Production of triterpenoids in the engineered strains overexpressing heterogenous GCSs[The same letters indicate no significant difference at the 95% confidence level (P < 0.05)]
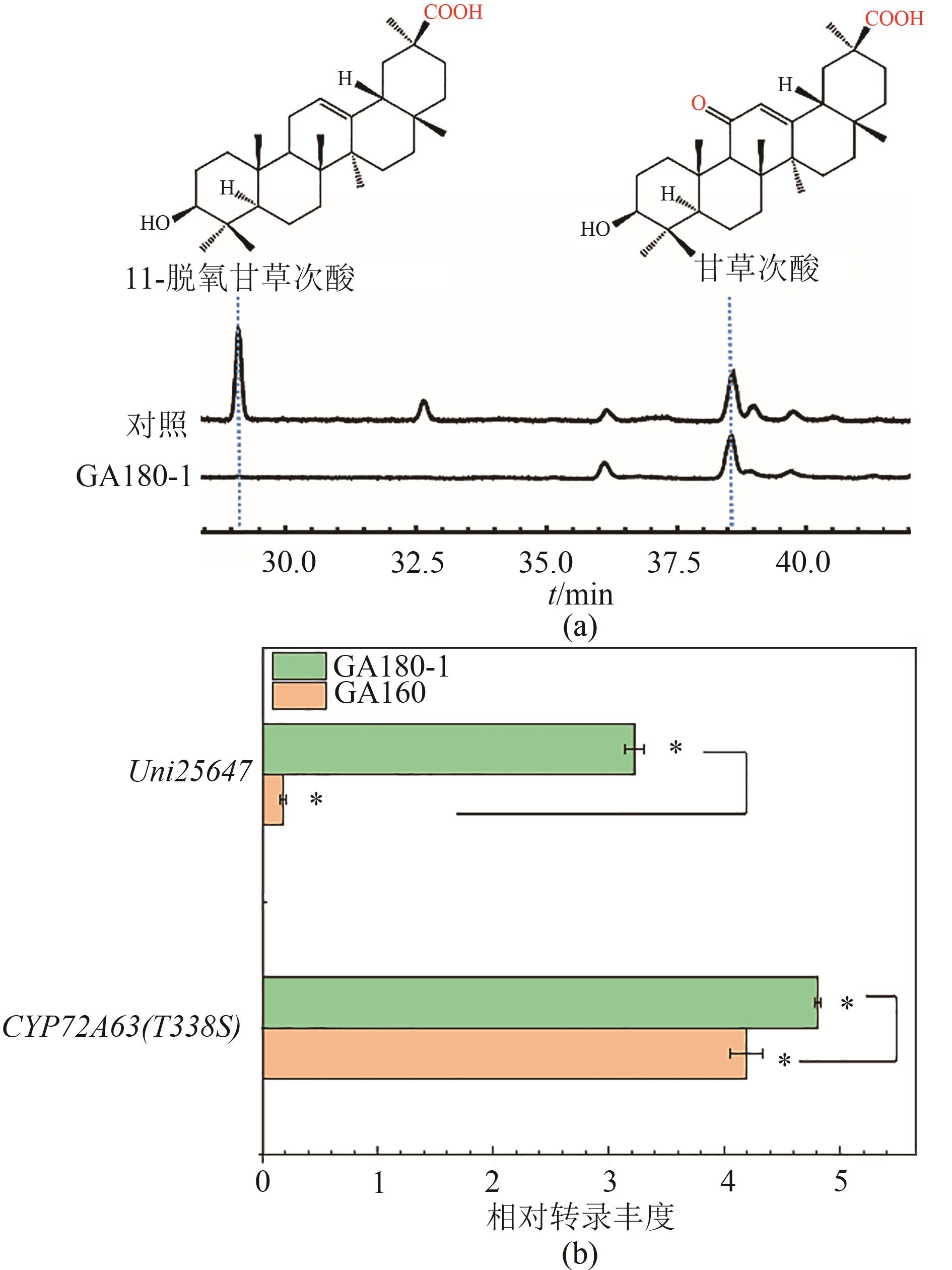
Fig. 6 Product profile (a) and the relative transcription abundance of P450 enzymes after up-regulating Uni25647 expression (b)[Asterisk (*) indicates significant difference at 95% confidence level (P < 0.05)]
| 1 | CHEMLER J A, KOFFAS M A. Metabolic engineering for plant natural product biosynthesis in microbes[J]. Current Opinion in Biotechnology, 2008, 19(6): 597-605. |
| 2 | SCHLÄGER S, DRÄGER B. Exploiting plant alkaloids[J]. Current Opinion in Biotechnology, 2016, 37: 155-164. |
| 3 | CHANG M C, EACHUS R A, TRIEU W, et al. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s[J]. Nature Chemical Biology, 2007, 3(5): 274-277. |
| 4 | GRANDNER J M, CACHO R A, TANG Y, et al. Mechanism of the P450-catalyzed oxidative cyclization in the biosynthesis of griseofulvin[J]. ACS Catalysis, 2016, 6(7): 4506-4511. |
| 5 | IGNEA C, ATHANASAKOGLOU A, IOANNOU E, et al. Carnosic acid biosynthesis elucidated by a synthetic biology platform[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(13): 3681-3686. |
| 6 | PARK H, PARK G, JEON W, et al. Whole-cell biocatalysis using cytochrome P450 monooxygenases for biotransformation of sustainable bioresources (fatty acids, fatty alkanes, and aromatic amino acids)[J]. Biotechnology Advances, 2020, 40: 107504. |
| 7 | NABAVI S M, ŠAMEC D, TOMCZYK M, et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering[J]. Biotechnology Advances, 2020, 38: 107316. |
| 8 | SEKI H, SAWAI S, OHYAMA K, et al. Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin[J]. The Plant Cell, 2011, 23(11): 4112-4123. |
| 9 | SEKI H, OHYAMA K, SAWAI S, et al. Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(37): 14204-14209. |
| 10 | DENISOV I G, MAKRIS T M, SLIGAR S G, et al. Structure and chemistry of cytochrome P450[J]. Chemical Reviews, 2005, 105(6): 2253-2277. |
| 11 | CHEN J, FAN F, QU G, et al. Identification of Absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone[J]. Metabolic Engineering, 2020, 57: 31-42. |
| 12 | SCHELER U, BRANDT W, PORZEL A, et al. Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast[J]. Nature Communications, 2016, 7: 12942. |
| 13 | CHEN W, FISHER M J, LEUNG A, et al. Oxidative diversification of steroids by nature-inspired scanning glycine mutagenesis of P450BM3 (CYP102A1)[J]. ACS Catalysis, 2020, 10(15): 8334-8343. |
| 14 | LI Y, WONG L L. Multi-functional oxidase activity of CYP102A1 (P450BM3) in the oxidation of quinolines and tetrahydroquinolines[J]. Angewandte Chemie International Edition, 2019, 58(28): 9551-9555. |
| 15 | LE-HUU P, HEIDT T, CLAASEN B, et al. Chemo-, regio-, and stereoselective oxidation of the monocyclic diterpenoid β-cembrenediol by P450 BM3[J]. ACS Catalysis, 2015, 5(3): 1772-1780. |
| 16 | URLACHER V B, GIRHARD M. Cytochrome P450 monooxygenases in biotechnology and synthetic biology[J]. Trends in Biotechnology, 2019, 37(8): 882-897. |
| 17 | BRANDENBERG O F, FASAN R, ARNOLD F H. Exploiting and engineering hemoproteins for abiological carbene and nitrene transfer reactions[J]. Current Opinion in Biotechnology, 2017, 47: 102-11. |
| 18 | ZHANG R K, HUANG X, ARNOLD F H. Selective CH bond functionalization with engineered heme proteins: new tools to generate complexity[J]. Current Opinion in Chemical Biology, 2019, 49: 67-75. |
| 19 | BULLER A R, ROYE P VAN, CAHN J K B, et al. Directed evolution mimics allosteric activation by stepwise tuning of the conformational ensemble[J]. Journal of the American Chemical Society, 2018, 140(23): 7256-7266. |
| 20 | SUN W, XUE H, LIU H, et al. Controlling chemo- and regioselectivity of a plant P450 in yeast cell toward rare licorice triterpenoid biosynthesis[J]. ACS Catalysis, 2020: 4253-4260. |
| 21 | 杨超凡,姜玉超,桑茉莉,等. 还原伴侣对细胞色素 P450 酶 MycG 的功能调控研究[J]. 合成生物学, 2021. doi: 10.12211/2096-8280.2021-053 . |
| YANG C F, JIANG Y C, SANG M L, et al. Studies on the functional modulating effects of redox partners on the cytochrome P450 enzyme MycG[J]. Synthetic Biology Journal, 2021. doi: 10.12211/2096-8280.2021-053 . | |
| 22 | KROGH A, LARSSON B, HEIJNE G VON, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes[J]. Journal of Molecular Biology, 2001, 305(3): 567-580. |
| 23 | ZHANG L, REN S, LIU X, et al. Mining of UDP-glucosyltrfansferases in licorice for controllable glycosylation of pentacyclic triterpenoids[J]. Biotechnology and Bioengineering, 2020, 117(12): 3651-3663. |
| 24 | ZHAO Y J, LI C. Biosynthesis of plant triterpenoid saponins in microbial cell factories[J]. Journal of Agricultural and Food Chemistry, 2018, 66(46): 12155-12165. |
| 25 | LIU H, FAN J, WANG C, et al. Enhanced β-amyrin synthesis in Saccharomyces cerevisiae by coupling an optimal acetyl-CoA supply pathway[J]. Journal of Agricultural and Food Chemistry, 2019, 67(13): 3723-3732. |
| 26 | ESPINOZA R V, SHERMAN D H. Exploring the molecular basis for selective C-H functionalization in plant P450s[J]. Synthetic and Systems Biotechnology, 2020, 5(2): 97-98. |
| 27 | ZHAO F, BAI P, LIU T, et al. Optimization of a cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2016, 113(8): 1787-1795. |
| 28 | MAMODE CASSIM A, GOUGUET P, GRONNIER J, et al. Plant lipids: Key players of plasma membrane organization and function[J]. Progress in Lipid Research, 2019, 73: 1-27. |
| 29 | BRIGNAC-HUBER L M, PARK J W, REED J R, et al. Cytochrome P450 organization and function are modulated by endoplasmic reticulum phospholipid heterogeneity[J]. Drug Metabolism and Disposition: the Biological Fate of Chemicals, 2016, 44(12): 1859-1866. |
| 30 | SEZGIN E, LEVENTAL I, MAYOR S, et al. The mystery of membrane organization: composition, regulation and roles of lipid rafts[J]. Nature Reviews Molecular Cell Biology, 2017, 18(6): 361-374. |
| 31 | HENRY S A, KOHLWEIN S D, CARMAN G M. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae [J]. Genetics, 2012, 190(2): 317-349. |
| 32 | ZHANG G, CAO Q, LIU J, et al. Refactoring β-amyrin synthesis in Saccharomyces cerevisiae [J]. AIChE Journal, 2015, 61(10): 3172-3179. |
| 33 | XIE Z X, LI B Z, MITCHELL L A, et al. "Perfect" designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 34 | PARISI L R, SOWLATI-HASHJIN S, BERHANE I A, et al. Membrane disruption by very long chain fatty acids during necroptosis[J]. ACS Chemical Biology, 2019, 14(10): 2286-2294. |
| 35 | DICKSON R C. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast[J]. Journal of Lipid Research, 2008, 49(5): 909-921. |
| 36 | MASHIMA R, OKUYAMA T, OHIRA M. Biosynthesis of long chain base in sphingolipids in animals, plants and fungi[J]. Future Science OA, 2019, 6(1): FSO434. |
| 37 | RAMIREZ-GAONA M, MARCU A, PON A, et al. YMDB 2.0: a significantly expanded version of the yeast metabolome database[J]. Nucleic Acids Research, 2017, 45(D1): D440-D445. |
| [1] | CHENG Xiaolei, LIU Tiangang, TAO Hui. Recent research progress in non-canonical biosynthesis of terpenoids [J]. Synthetic Biology Journal, 2024, 5(5): 1050-1071. |
| [2] | PAN Yingjia, XIA Siyang, DONG Chang, CAI Jin, LIAN Jiazhang. Mutator-driven continuous genome evolution of Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2023, 4(1): 225-240. |
| [3] | YANG Chaofan, JIANG Yuchao, SANG Moli, LI Shengying, ZHANG Wei. Studies on the functional modulating effect of redox partners on the cytochrome P450 enzyme MycG [J]. Synthetic Biology Journal, 2022, 3(3): 587-601. |
| [4] | LYU Jianming, ZHAO Huan, HU Dan, GAO Hao. Biosynthesis of alkyne moiety in natural products and application of alkyne biosynthetic machineries [J]. Synthetic Biology Journal, 2021, 2(5): 734-750. |
| [5] | LI Xiaodong, YANG Chengshuai, WANG Pingping, YAN Xing, ZHOU Zhihua. Production of sesquiterpenoids α-neoclovene and β-caryophyllene by engineered Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2021, 2(5): 792-803. |
| [6] | Yi LI, Zhenquan LIN, Zihe LIU. Advances in yeast based adaptive laboratory evolution [J]. Synthetic Biology Journal, 2021, 2(2): 287-301. |
| [7] | Yue SHENG, Genlin ZHANG. Yeast terminator engineering: from mechanism exploration to artificial design [J]. Synthetic Biology Journal, 2020, 1(6): 709-721. |
| [8] | Siyang XIA, Lihong JIANG, Jin CAI, Lei HUANG, Zhinan XU, Jiazhang LIAN. Advances in genome evolution of Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2020, 1(5): 556-569. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||