Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (5): 837-849.DOI: 10.12211/2096-8280.2021-024
Genome mining for novel natural products in Sorangium cellulosum So0157-2 by heterologous expression
ZHOU Haibo, SHEN Qiyao, CHEN Hanna, WANG Zongjie, LI Yuezhong, ZHANG Youming, BIAN Xiaoying
- State Key Laboratory of Microbial Technology,Shandong University,Qingdao 266237,Shandong,China
-
Received:2021-02-20Revised:2021-04-06Online:2021-11-19Published:2021-10-31 -
Contact:ZHANG Youming, BIAN Xiaoying
利用异源表达挖掘纤维堆囊菌So0157-2的新型天然产物
周海波, 申琪瑶, 陈汉娜, 王宗杰, 李越中, 张友明, 卞小莹
- 山东大学,微生物技术国家重点实验室,山东 青岛 266237
-
通讯作者:张友明,卞小莹 -
作者简介:周海波 (1987—),男,博士,助理研究员。研究方向为微生物次级代谢产物的发现及其生物合成途径解析。E-mail:haibozhou@sdu.edu.cn张友明 (1964—),男,博士生导师,教授。研究方向为基因组编辑与合成生物学等。E-mail:zhangyouminng@sdu.edu.cn卞小莹 (1983—),男,博士生导师,教授。研究方向为微生物天然产物合成生物学等。E-mail:bianxiaoying@sdu.edu.cn -
基金资助:国家重点研发计划(2019YFA0905700);国家自然科学基金(32070060);山东省自然科学基金(ZR2020QH345)
CLC Number:
Cite this article
ZHOU Haibo, SHEN Qiyao, CHEN Hanna, WANG Zongjie, LI Yuezhong, ZHANG Youming, BIAN Xiaoying. Genome mining for novel natural products in Sorangium cellulosum So0157-2 by heterologous expression[J]. Synthetic Biology Journal, 2021, 2(5): 837-849.
周海波, 申琪瑶, 陈汉娜, 王宗杰, 李越中, 张友明, 卞小莹. 利用异源表达挖掘纤维堆囊菌So0157-2的新型天然产物[J]. 合成生物学, 2021, 2(5): 837-849.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-024
| Strains/Plasmids | Description | Sources |
|---|---|---|
| Strains | [ | |
| Escherichia coli GB05-dir | GB2005, araC-PBAD-ETgA; recE, recT, redγ, and recA regulated by arabinose-induced promoter are inserted at ybcC locus | |
| E. coli GB05-red | GB2005, araC-PBAD-αβγA; redα, redβ, redγ, and recA regulated by arabinose-induced promoter are integrated at ybcC locus | [ |
| S. cellulosum So0157-2 | wild type harboring BGC18 | [ |
| S. brevitalea DSM 7029 | Burkholderiales strain DSM 7029, [Polyangium] brachysporum DSM 7029 (K481-B101; ATCC 53080) | [ |
| DSM7029:Pkm-BGC18 | Pkm-BGC18 was inserted into genome of Schlegelella brevitalea DSM 7029, kmR | This work |
| Plasmids | ||
| p15A-cm-tetR-tetO-hyg-ccdB | direct cloning vector, p15A replicon, containing a tetracycline inducible promoter PtetO, cmR/hygR | [ |
| pR6K-oriT-TnpA-IR-km | R6K replicon, containing MycoMar transposase gene (tnpA) and conjugation element oriT, kmR | [ |
| p15A-cm-BGC18 | BGC18 was cloned into cloning vector, cmR | This work |
| p15A-oriT-IR-Pkm-BGC18 | Expression vector of BGC18 with Pkm was inserted into upstream of the first core gene of BGC18, containing transposon elements oriT-tnpA-IR, kmR | This work |
Tab. 1 Strains and plasmids used in this study
| Strains/Plasmids | Description | Sources |
|---|---|---|
| Strains | [ | |
| Escherichia coli GB05-dir | GB2005, araC-PBAD-ETgA; recE, recT, redγ, and recA regulated by arabinose-induced promoter are inserted at ybcC locus | |
| E. coli GB05-red | GB2005, araC-PBAD-αβγA; redα, redβ, redγ, and recA regulated by arabinose-induced promoter are integrated at ybcC locus | [ |
| S. cellulosum So0157-2 | wild type harboring BGC18 | [ |
| S. brevitalea DSM 7029 | Burkholderiales strain DSM 7029, [Polyangium] brachysporum DSM 7029 (K481-B101; ATCC 53080) | [ |
| DSM7029:Pkm-BGC18 | Pkm-BGC18 was inserted into genome of Schlegelella brevitalea DSM 7029, kmR | This work |
| Plasmids | ||
| p15A-cm-tetR-tetO-hyg-ccdB | direct cloning vector, p15A replicon, containing a tetracycline inducible promoter PtetO, cmR/hygR | [ |
| pR6K-oriT-TnpA-IR-km | R6K replicon, containing MycoMar transposase gene (tnpA) and conjugation element oriT, kmR | [ |
| p15A-cm-BGC18 | BGC18 was cloned into cloning vector, cmR | This work |
| p15A-oriT-IR-Pkm-BGC18 | Expression vector of BGC18 with Pkm was inserted into upstream of the first core gene of BGC18, containing transposon elements oriT-tnpA-IR, kmR | This work |
| Primers | Sequences (5′-3′) |
|---|---|
| BGC18-HAF | cagtatggccgatcggggtgtcagcggtcaacacgcgagctcgctcgctctgtggcggcgctctcatggtgcAACGCTCTCTACTAGAGTCA |
| BGC18-HAR | gacgtgaccgggtatcgctaggtccacccaccaggagtccggccagggatccggagagtgagagttcaacgcGGGTCTTAAGACGTCGATATCT |
| IR-Pkm-C18-HAF1 | gcctgcgatcgtaccattacgtatttttgcgcgagcggaacggtatgcagGCTGATCTTCAGATCCTCTAC |
| IR-Pkm-C18-HAR1 | tcgagccgcaaggcgcattcttgctccagagagagcggcatagtacccaacctcctTCAGAAGAACTCGTCAAGAAG |
| detect-C18 in 7029-F1 | GATGGGTTATCAGGACTACGC |
| detect-C18 in 7029-R1 | CGAGGAGCCTGTAGAACGCGT |
| detect-C18 in 7029-F2 | TCGACGATCAGGTGAAGATCCA |
| detect-C18 in 7029-R2 | AGCTCTCCATAGGTGAGCGAA |
| detect-C18 in 7029-F3 | TGAAGATCCGCGGCTATCGCA |
| detect-C18 in 7029-R3 | CGGGAACTGGAACAGGTCCAT |
| detect-C18 in 7029-F4 | TTCTTCGTCAACGCCGCGCC |
| detect-C18 in 7029-R4 | GCGATAGGCGTGCACCGTGC |
| detect-C18 in 7029-F5 | TCACGCTGCCGCAAGCACTC |
| detect-C18 in 7029-R5 | GCTCTGCGAAGGACGTCCTC |
Tab. 2 Primers used in this study
| Primers | Sequences (5′-3′) |
|---|---|
| BGC18-HAF | cagtatggccgatcggggtgtcagcggtcaacacgcgagctcgctcgctctgtggcggcgctctcatggtgcAACGCTCTCTACTAGAGTCA |
| BGC18-HAR | gacgtgaccgggtatcgctaggtccacccaccaggagtccggccagggatccggagagtgagagttcaacgcGGGTCTTAAGACGTCGATATCT |
| IR-Pkm-C18-HAF1 | gcctgcgatcgtaccattacgtatttttgcgcgagcggaacggtatgcagGCTGATCTTCAGATCCTCTAC |
| IR-Pkm-C18-HAR1 | tcgagccgcaaggcgcattcttgctccagagagagcggcatagtacccaacctcctTCAGAAGAACTCGTCAAGAAG |
| detect-C18 in 7029-F1 | GATGGGTTATCAGGACTACGC |
| detect-C18 in 7029-R1 | CGAGGAGCCTGTAGAACGCGT |
| detect-C18 in 7029-F2 | TCGACGATCAGGTGAAGATCCA |
| detect-C18 in 7029-R2 | AGCTCTCCATAGGTGAGCGAA |
| detect-C18 in 7029-F3 | TGAAGATCCGCGGCTATCGCA |
| detect-C18 in 7029-R3 | CGGGAACTGGAACAGGTCCAT |
| detect-C18 in 7029-F4 | TTCTTCGTCAACGCCGCGCC |
| detect-C18 in 7029-R4 | GCGATAGGCGTGCACCGTGC |
| detect-C18 in 7029-F5 | TCACGCTGCCGCAAGCACTC |
| detect-C18 in 7029-R5 | GCTCTGCGAAGGACGTCCTC |
| Gene Clusters | Type | Similar known cluster | Similarity |
|---|---|---|---|
BGC1 BGC2 BGC3 BGC4 BGC5 BGC6 BGC7 BGC8 BGC9 BGC10 BGC11 BGC12 BGC13 BGC14 BGC15 BGC16 BGC17 BGC18 BGC19 BGC20 BGC21 BGC22 BGC23 BGC24 BGC25 BGC26 BGC27 BGC28 BGC29 BGC30 BGC31 BGC32 BGC33 BGC34 BGC35 | RiPP-like NRPS Thioamitides NRPS-PKS NRPS-PKS hglE-KS RiPP-like RiPP-like Indole T3PKS NRPS-T1PKS Terpene T1PKS NRPS RiPP-like thioamitides NRPS NRPS-T1PKS NRPS-T1PKS hglE-KS NRPS-T1PKS NRPS-T1PKS arylpolyene LAP, RRE-containing RiPP-like RiPP-like T1PKS T1PKS microbiridin phosphonate NRPS RiPP-like NRPS-T1PKS terpene NRPS | — — — pyxipyrrolone A/B kirromycin — — — — alkylpyrone-407/393 disorazol A geosmin pallasoren — — — myxochelin A/B — Epothilone — — Hapalosin N-tetradecanoyl tyrosine — — — — — microviridin J — coelibactin — crochelin A eremophikene — | — — — 11% 5% — — — — 6% 23% 100% 50% — — — 50% — 100% — — 40% 6% — — — — — 66% — 45% — 26% 100% — |
Tab. 3 Biosynthetic gene clusters in the genome of so0157-2 predicted by antiSMASH
| Gene Clusters | Type | Similar known cluster | Similarity |
|---|---|---|---|
BGC1 BGC2 BGC3 BGC4 BGC5 BGC6 BGC7 BGC8 BGC9 BGC10 BGC11 BGC12 BGC13 BGC14 BGC15 BGC16 BGC17 BGC18 BGC19 BGC20 BGC21 BGC22 BGC23 BGC24 BGC25 BGC26 BGC27 BGC28 BGC29 BGC30 BGC31 BGC32 BGC33 BGC34 BGC35 | RiPP-like NRPS Thioamitides NRPS-PKS NRPS-PKS hglE-KS RiPP-like RiPP-like Indole T3PKS NRPS-T1PKS Terpene T1PKS NRPS RiPP-like thioamitides NRPS NRPS-T1PKS NRPS-T1PKS hglE-KS NRPS-T1PKS NRPS-T1PKS arylpolyene LAP, RRE-containing RiPP-like RiPP-like T1PKS T1PKS microbiridin phosphonate NRPS RiPP-like NRPS-T1PKS terpene NRPS | — — — pyxipyrrolone A/B kirromycin — — — — alkylpyrone-407/393 disorazol A geosmin pallasoren — — — myxochelin A/B — Epothilone — — Hapalosin N-tetradecanoyl tyrosine — — — — — microviridin J — coelibactin — crochelin A eremophikene — | — — — 11% 5% — — — — 6% 23% 100% 50% — — — 50% — 100% — — 40% 6% — — — — — 66% — 45% — 26% 100% — |
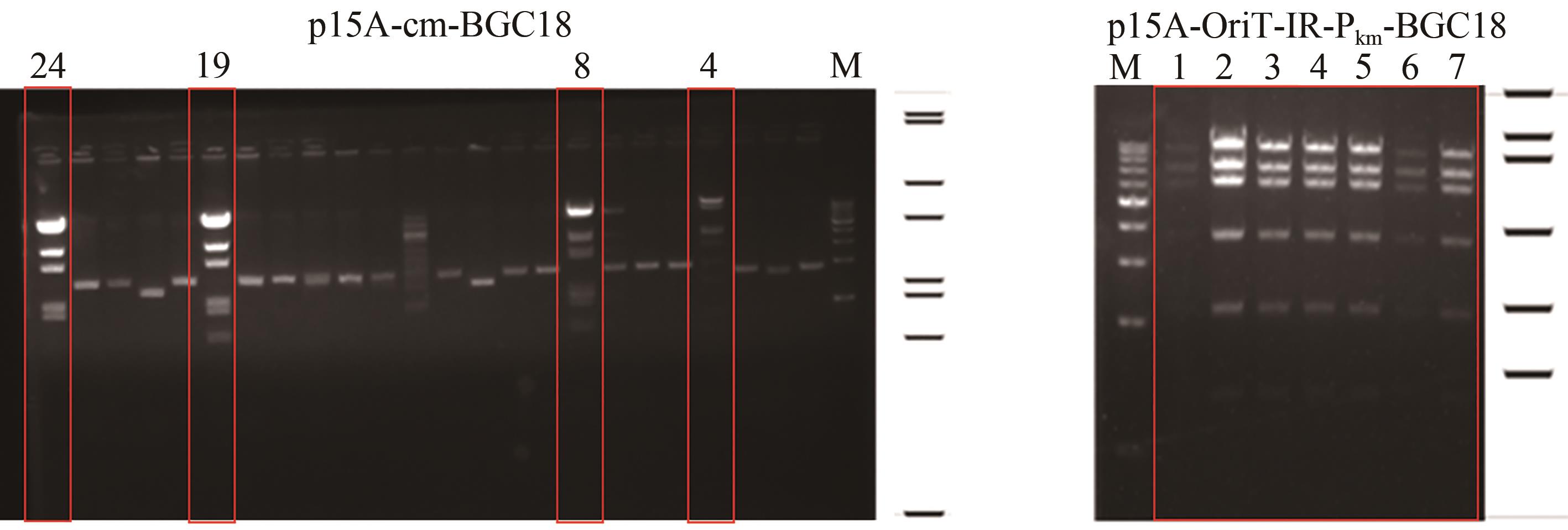
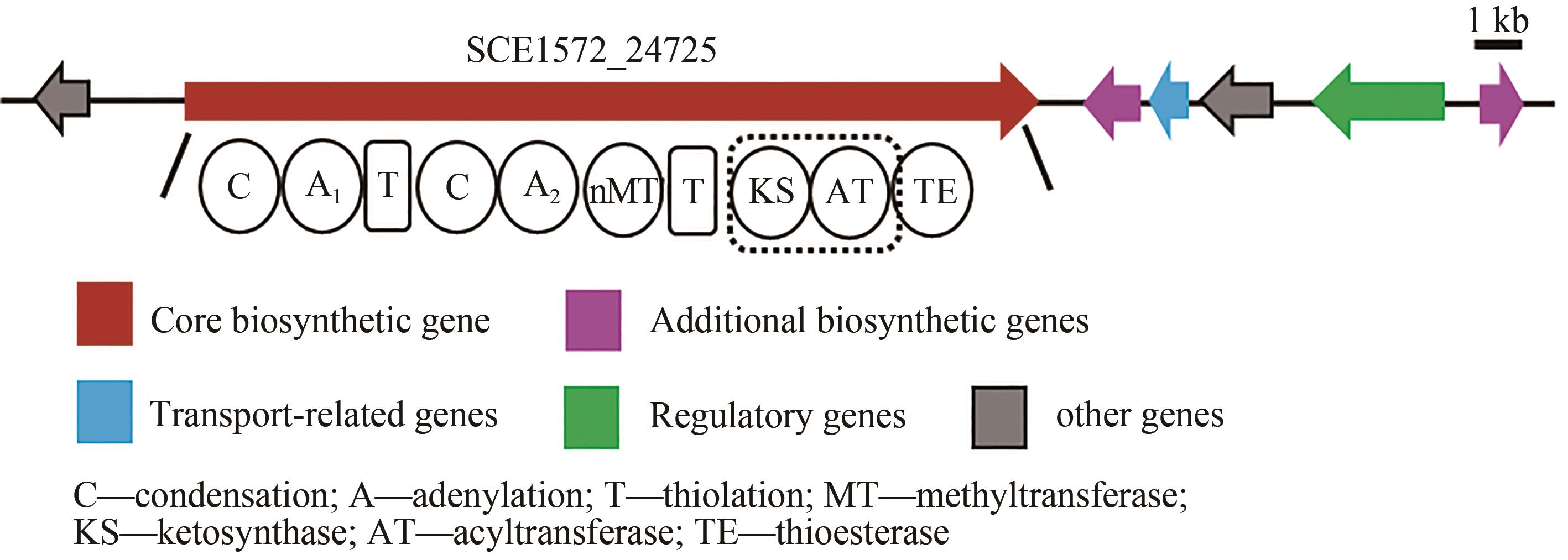
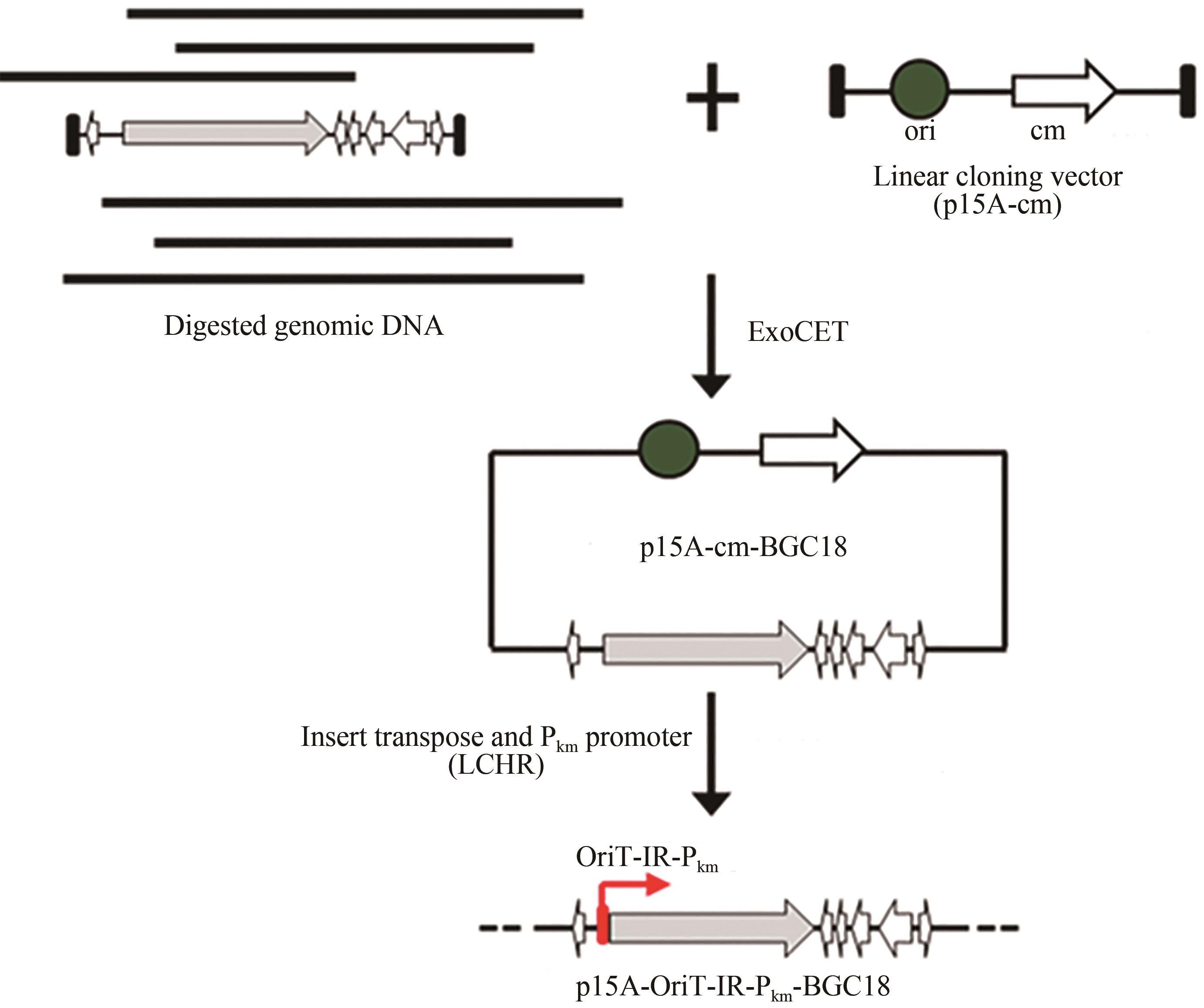
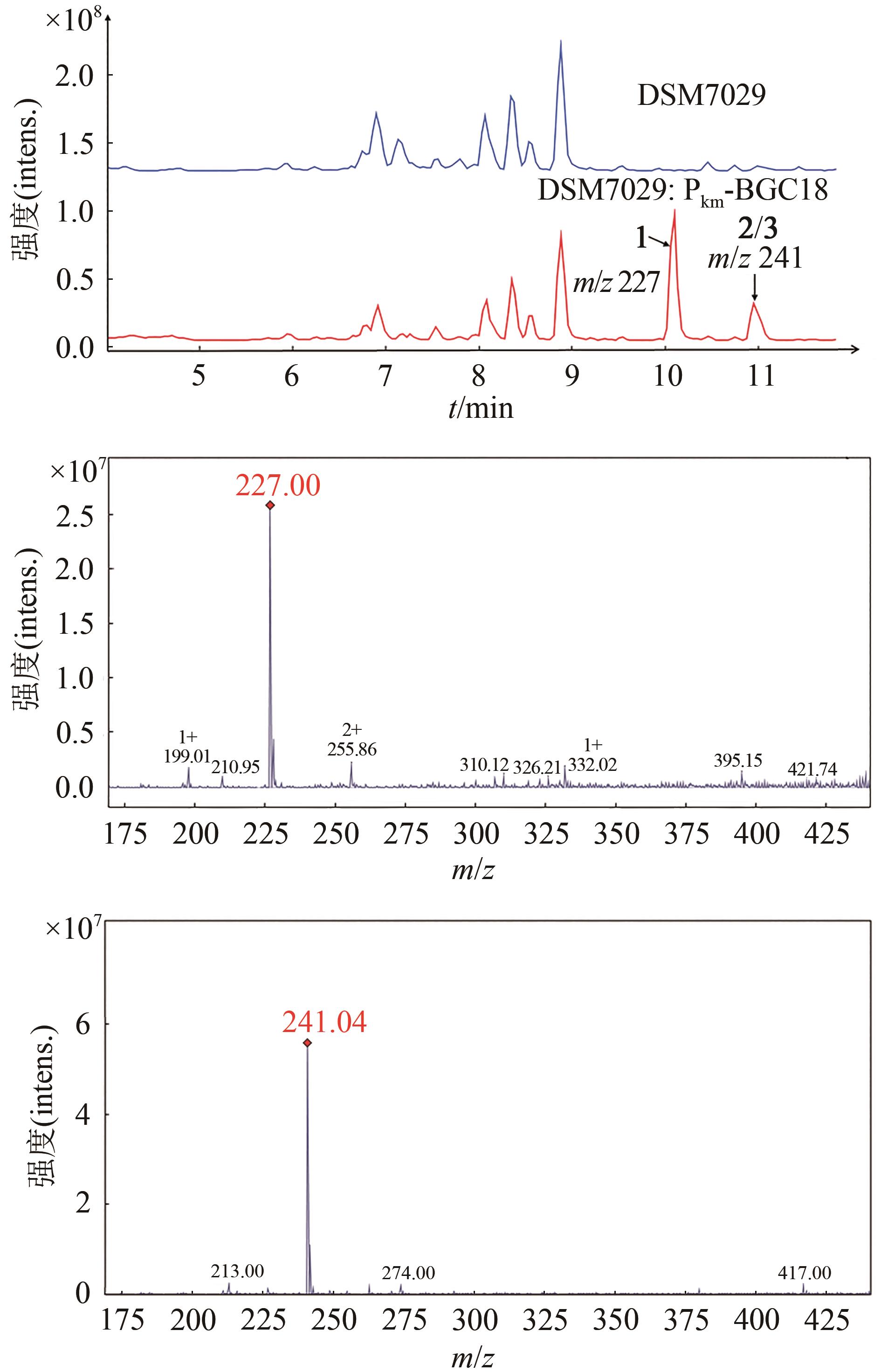
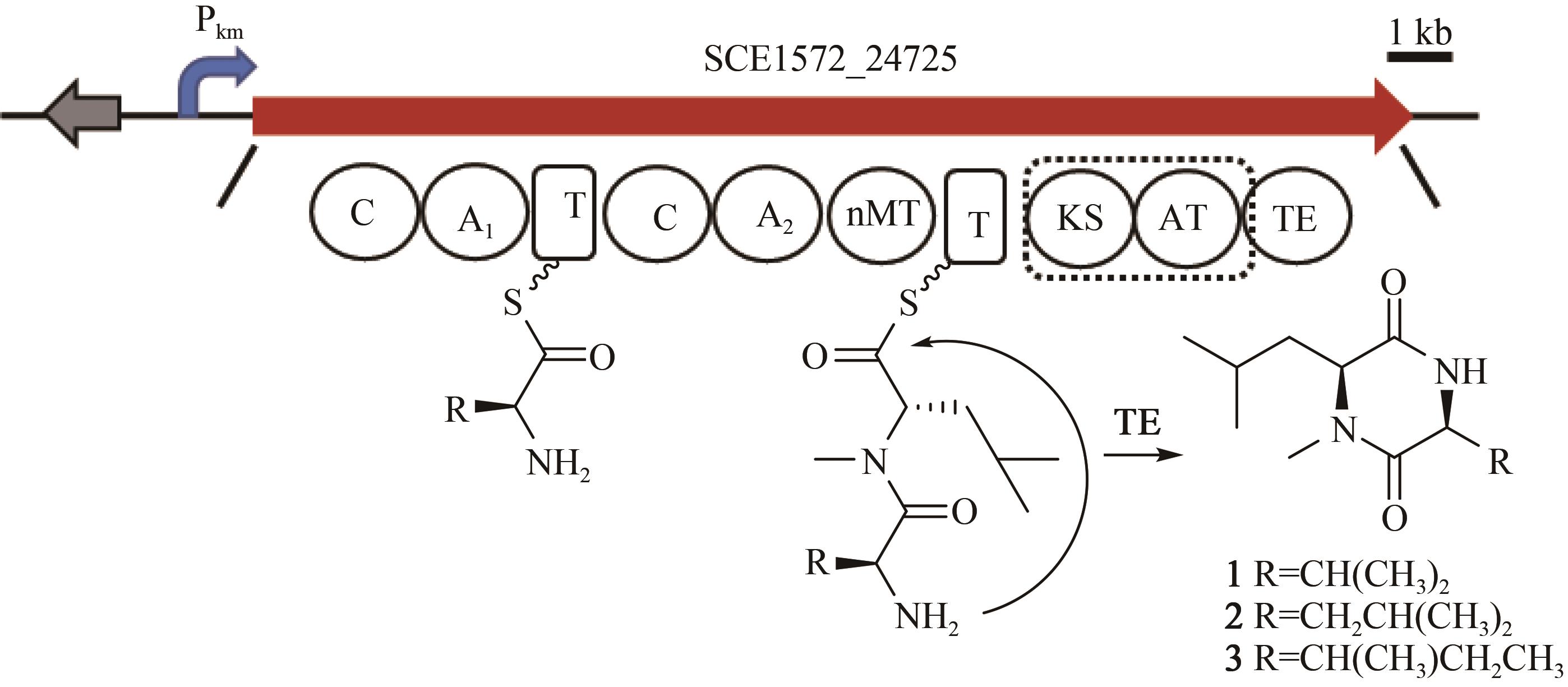
Fig. 3 a: Restriction analysis of the recombinant plasmids p15A-cm-BGC18 by MscⅠ(a) and p15A-OriT-IR-Pkm-BGC18 by ApaLⅠ(b)(Red box indicates right recombinant plasmids)
| No | 1 | 2 | 3 | ||
|---|---|---|---|---|---|
| δC | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | ||
| N-Me-Leu | 1 2 3 4 5 6 N-Me | 167.6, C 59.2, CH 42.7, CH2 25.1, CH 23.1, CH3 21.8, CH3 32.1, CH3 | 3.78, dd (4.5, 8.2) 1.58, m 1.87, m 0.89, d (6.7) 0.93, d (6.7) 2.82, s | 3.79, t (6.7) 1.59, t (7.0) 1.79, m 0.90, d (6.5) 0.93, d (6.5) 2.82, s | 3.78, dd (4.5, 8.1) 1.58, m 1.87, m 0.89, d (6.5) 0.93, d (6.5) 2.82, s |
| Val/Leu/Ile | 1 2 3 4 5 6 NH | 165.1, C 60.1, CH 32.6, CH 19.2, CH3 18.1, CH3 | 3.55, dd (3.6, 5.2) 2.01, m 0.92, d (6.8) 0.84, d (6.8) 8.24, d (2.4) | 3.73, dt (3.9, 8.7) 1.52, m 1.45, m 1.79, m 0.86, d (6.5) 0.90, d (6.5) 8.39, d (2.6) | 3.61, dd (3.8, 4.8) 1.73, m 1.43, m 1.11, m 0.84, t (6.5) 0.89, d (6.5) 8.23, d (1.6) |
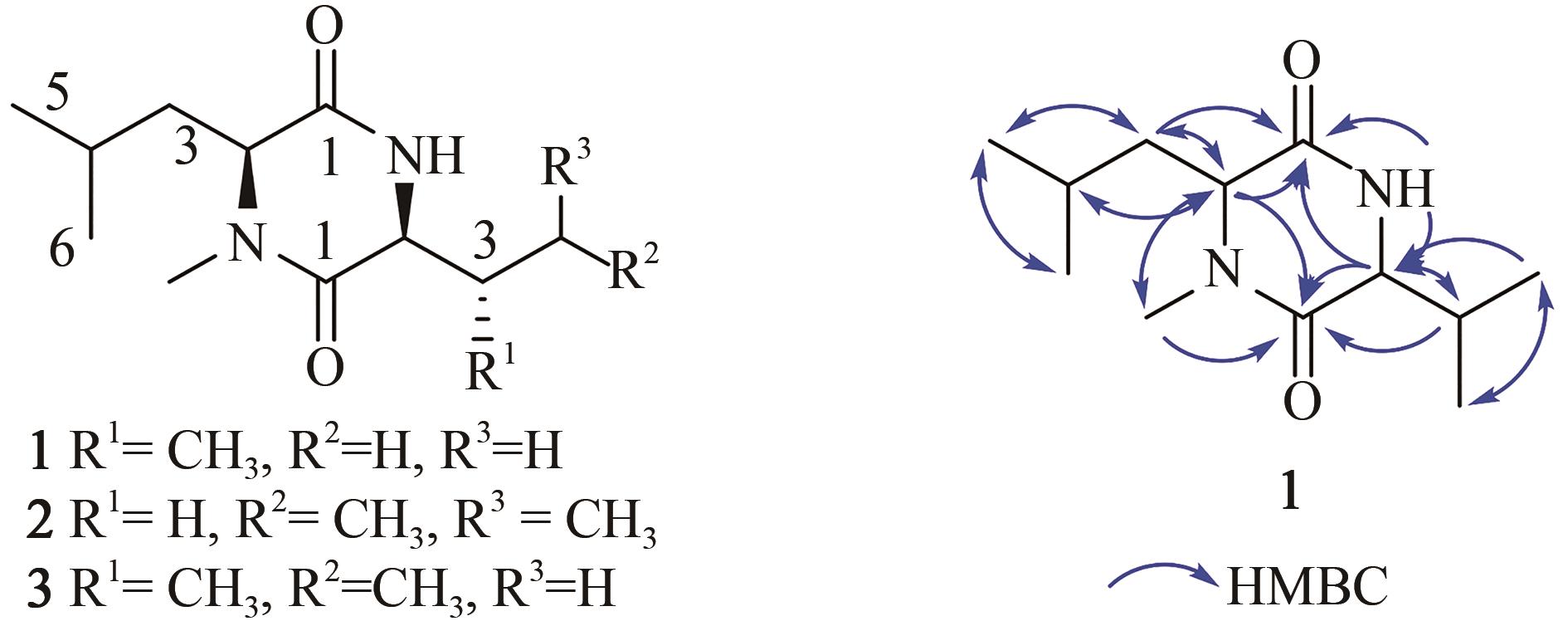
Tab. 4 1H (500 MHz) and 13C (125 MHz) data of 1~3 in DMSO-d6
| No | 1 | 2 | 3 | ||
|---|---|---|---|---|---|
| δC | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | ||
| N-Me-Leu | 1 2 3 4 5 6 N-Me | 167.6, C 59.2, CH 42.7, CH2 25.1, CH 23.1, CH3 21.8, CH3 32.1, CH3 | 3.78, dd (4.5, 8.2) 1.58, m 1.87, m 0.89, d (6.7) 0.93, d (6.7) 2.82, s | 3.79, t (6.7) 1.59, t (7.0) 1.79, m 0.90, d (6.5) 0.93, d (6.5) 2.82, s | 3.78, dd (4.5, 8.1) 1.58, m 1.87, m 0.89, d (6.5) 0.93, d (6.5) 2.82, s |
| Val/Leu/Ile | 1 2 3 4 5 6 NH | 165.1, C 60.1, CH 32.6, CH 19.2, CH3 18.1, CH3 | 3.55, dd (3.6, 5.2) 2.01, m 0.92, d (6.8) 0.84, d (6.8) 8.24, d (2.4) | 3.73, dt (3.9, 8.7) 1.52, m 1.45, m 1.79, m 0.86, d (6.5) 0.90, d (6.5) 8.39, d (2.6) | 3.61, dd (3.8, 4.8) 1.73, m 1.43, m 1.11, m 0.84, t (6.5) 0.89, d (6.5) 8.23, d (1.6) |
| Amino acid | Configuration | Retention time | |||
|---|---|---|---|---|---|
| Standard | 1 | 2 | 3 | ||
| N-Me-L-Leu | L | 12.9 | 12.9 | 12.9 | 12.9 |
| D | 13.2 | ||||
| Val | L | 11.6 | 11.6 | ||
| D | 12.5 | ||||
| Leu | L | 29.7 | 29.7 | ||
| D | 33.1 | ||||
| Ile | L | 29.1 | 29.1 | ||
| D | 32.5 | ||||
| allo-Ile | L | 29.2 | |||
| D | 32.6 | ||||
Tab. 5 Retention time of amino acids derivatized with Marfey′s reagent
| Amino acid | Configuration | Retention time | |||
|---|---|---|---|---|---|
| Standard | 1 | 2 | 3 | ||
| N-Me-L-Leu | L | 12.9 | 12.9 | 12.9 | 12.9 |
| D | 13.2 | ||||
| Val | L | 11.6 | 11.6 | ||
| D | 12.5 | ||||
| Leu | L | 29.7 | 29.7 | ||
| D | 33.1 | ||||
| Ile | L | 29.1 | 29.1 | ||
| D | 32.5 | ||||
| allo-Ile | L | 29.2 | |||
| D | 32.6 | ||||
| 1 | HERRMANN J, FAYAD A A, MÜLLER R. Natural products from myxobacteria: novel metabolites and bioactivities[J]. Natural Product Reports, 2017, 34(2): 135-160. |
| 2 | WEISSMAN K J, MÜLLER R. Myxobacterial secondary metabolites: bioactivities and modes-of-action[J]. Natural Product Reports, 2010, 27(9): 1276-1295. |
| 3 | WENZEL S C, MÜLLER R. Myxobacteria—'microbial factories' for the production of bioactive secondary metabolites[J]. Molecular Biosystems, 2009, 5(6): 567-574. |
| 4 | SCHÄBERLE T F, LOHR F, SCHMITZ A, et al. Antibiotics from myxobacteria[J]. Natural Product Reports, 2014, 31(7): 953-972. |
| 5 | 刘冰花. 黏细菌生物活性物质的研究[J]. 成都大学学报(自然科学版), 2011, 30(2): 108-113. |
| LIU B H. Study of myxobacterial bioactive substances[J]. Journal of Chengdu University (Natural Science), 2011, 30(2): 108-113. | |
| 6 | 赵婷峰, 龚国利. 黏细菌: 天然的制药厂[J]. 生物技术通报, 2014(12): 40-46. |
| ZHAO T F, GONG G L. Myxobacteria: natural pharmaceutical factories[J]. Biotechnology Bulletin, 2014(12): 40-46. | |
| 7 | HAN K, LI Z F, PENG R, et al. Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu[J]. Scientific Reports, 2013, 3: 2101. |
| 8 | LI Z F, ZHU L P, GU J Y, et al. Isolation and characterisation of the epothilone gene cluster with flanks from high alkalotolerant strain Sorangium cellulosum (So0157-2)[J]. World Journal of Microbiology and Biotechnology, 2017, 33(7): 137. |
| 9 | 龚国利, 黄宇莹. 黏细菌纤维堆囊菌的基因组学研究现状[J]. 基因组学与应用生物学, 2014, 33(5): 1133-1138. |
| GONG G L, HUANG Y Y. Research status of genomics of myxobacteria Sorangium cellulosum [J]. Genomics and Applied Biology, 2014, 33(5): 1133-1138. | |
| 10 | ZHAO L, LI P F, LU C H, et al. Glycosylation and production characteristics of epothilones in alkali-tolerant Sorangium cellulosum strain So0157-2[J]. The Journal of Microbiology, 2010, 48(4): 438-444. |
| 11 | WANG J, ZHANG H, YING L, et al. Five new epothilone metabolites from Sorangium cellulosum strain So0157-2[J]. The Journal of Antibiotics, 2009, 62: 483-487. |
| 12 | ZHAO L, ZHAO B B, LU C H, et al. Two epothilones from Sorangium cellulosum strain So0157-2[J]. Chinese Journal of Natural Medicines, 2010, 8: 298-300. |
| 13 | WANG J D, JIANG N, ZHANG H, et al. New epothilone congeners from Sorangium cellulosum strain So0157-2[J]. Natural Product Research, 2011, 25(18): 1707-1712. |
| 14 | LU C, ZHAO L, LI Y, et al. Four natural epothilone derivatives isolated from Sorangium cellulosum strain So0157-2[J]. Journal of Chinese Pharmaceutical Sciences, 2013, 22(1): 28-31. |
| 15 | ZHANG Y J, DENG A W, ZHANG H, et al. Epothilone O, a new member of this family from Sorangium cellulosum strain So0157-2[J]. The Journal of Antibiotics, 2013, 66(5): 285-286. |
| 16 | HUO L, HUG J J, FU C, et al. Heterologous expression of bacterial natural product biosynthetic pathways[J]. Natural Product Reports, 2019, 36(10): 1412-1436. |
| 17 | ONGLEY S E, BIAN X, NEILAN B A, et al. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways[J]. Natural Product Reports, 2013, 30: 1121-1138. |
| 18 | ZHANG J J, TANG X Y, MOORE B S. Genetic platforms for heterologous expression of microbial natural products[J]. Natural Product Reports, 2019, 36(9): 1313-1332. |
| 19 | LUO Y, ENGHIAD B, ZHAO H. New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters[J]. Natural Product Reports, 2016, 33(2): 174-182. |
| 20 | LUO Y, LI B Z, LIU D, et al. Engineered biosynthesis of natural products in heterologous hosts[J]. Chemical Society Reviews, 2015, 44(15): 5265-5290. |
| 21 | FU J, BIAN X Y, HU S B, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting[J]. Nature Biotechnology, 2012, 30(5): 440-446. |
| 22 | WANG H L, LI Z, JIA R N, et al. ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes[J]. Nucleic Acids Research, 2018, 46(5): e28. |
| 23 | 郑文韬, 张友明, 卞小莹. Red/ET同源重组技术及其在微生物基因组挖掘中的应用进展[J]. 微生物学报, 2017, 57(11): 1735-1746. |
| ZHENG W T, ZHANG Y M, BIAN X Y. Advances in Red/ET recombineering and its application for microbial genome mining[J]. Acta Microbiologica Sinica, 2017, 57(11): 1735-1746. | |
| 24 | WANG H, BIAN X, XIA L, et al. Improved seamless mutagenesis by recombineering using ccdB for counterselection[J]. Nucleic Acids Research, 2014, 42(5): e37. |
| 25 | BIAN X, PLAZA A, YAN F, et al. Rational and efficient site-directed mutagenesis of adenylation domain alters relative yields of luminmide derivatives in vivo [J]. Biotechnology and Bioengineering, 2015, 112(7): 1343-1353. |
| 26 | WANG H L, LI Z, JIA R N, et al. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression[J]. Nature Protocols. 2016, 11(7): 1175-1190. |
| 27 | ABBASI M N, FU J, BIAN X, et al. Recombineering for genetic engineering of natural product biosynthetic pathways[J]. Trends in Biotechnology, 2020, 38(7): 715-728. |
| 28 | ZHONG L, DIAO X, ZHANG N, et al. Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis[J]. Nature Communications, 2021, 12(1): 296. |
| 29 | WATANABE K, OIKAWA H. Robust platform for de novo production of heterologous polyketides and nonribosomal peptides in Escherichia coli [J]. Organic & Biomolecular Chemistry, 2007, 5(4): 593-602. |
| 30 | 马紫卉, 李伟, 尹文兵. 真菌天然产物异源生产研究进展[J]. 微生物学报, 2016, 56(3): 429-440. |
| MA Z H, LI W, YIN W B. Progress in heterologous expression of fungal natural products-a review[J]. Acta Microbiologica Sinica, 2016, 56(3): 429-440. | |
| 31 | TSUNEMATSU Y, ISHIUCHI K, HOTTA K, et al. Yeast-based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products[J]. Natural Product Reports, 2013, 30(8): 1139-1149. |
| 32 | MYRONOVSKYI M, LUZHETSKYY A. Heterologous production of small molecules in the optimized Streptomyces hosts[J]. Natural Product Reports, 2019, 36(9): 1281-1294. |
| 33 | 王素英, 蒋雪, 张宏宇. 放线菌次级代谢产物合成基因簇异源表达体系的研究进展[J]. 科学技术与工程, 2019, 19(34): 9-16. |
| WANG S Y, JIANG X, ZHANG H Y. Advances in heterologous expression system of secondary metabolite synthesis gene clusters of actinomycetes[J]. Science Technology and Engineering, 2019, 19(34): 9-16. | |
| 34 | YU Y C, WANG H M, TANG B, et al. Reassembly of the biosynthetic gene cluster enables high epothilone yield in engineered Schlegelella brevitalea [J]. ACS Synthetic Biology, 2020, 9(8): 2009-2022. |
| 35 | WANG X, ZHOU H B, CHEN H N, et al. Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(18): E4255-E4263. |
| 36 | BIAN X Y, TANG B, YU Y C, et al. Heterologous production and yield improvement of epothilones in Burkholderiales strain DSM 7029[J]. ACS Chemical Biology, 2017, 12(7): 1805-1812. |
| 37 | OUYANG Q, WANG X, ZHANG N, et al. Promoter screening facilitates heterologous production of complex secondary metabolites in Burkholderiales strains[J]. ACS Synthetic Biology, 2020, 9(2): 457-460. |
| 38 | FU J, TEUCHER M., ANASTASSIADIS K, et al. Chapter eight-a recombineering pipeline to make conditional targeting constructs [J]. Methods in Enzymology, 2010, 477: 125-144. |
| 39 | TANG B, YU Y, LIANG J, et al. Reclassification of 'Polyangium brachysporum' DSM 7029 as Schlegelella brevitalea sp. nov[J]. International Journal of Systematic and Evolutionary Microbiology, 2019, 69(9): 2877-2883. |
| 40 | GIGLIO S, JIANG J Y, SAINT C P, et al. Isolation and characterization of the gene associated with Geosmin production in cyanobacteria[J]. Environmental Science & Technology, 2008, 42(21): 8027-8032. |
| 41 | SCHIFRIN A, LY T T, GÜNNEWICH N, et al. Characterization of the gene cluster CYP264B1-geoA from Sorangium cellulosum So ce56: Biosynthesis of (+)-eremophilene and its hydroxylation[J]. Chembiochem, 2015, 16(2): 337-344. |
| 42 | MIYANAGA A, KUDO F, EGUCHI T. Protein-protein interactions in polyketide synthase-nonribosomal peptide synthetase hybrid assembly lines[J]. Natural Product Reports, 2018, 35(11): 1185-1209. |
| 43 | WEN S J, HU T S, YAO Z J. Macrocyclization studies and total synthesis of cyclomarin C, an anti-inflammatory marine cyclopeptide[J]. Tetrahedron, 2005, 61(21): 4931-4938. |
| 44 | KATSUTA R, TOYODA M, YAJIMA A, et al. Synthesis and stereochemistry of JBIR-81, a peptide enamide derived from aspergilli [J]. Tetrahedron Letters. 2018, 59: 1010-1013. |
| 45 | CRYLE M J, BELL S G, SCHLICHTING I. Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: A cyclo-L-leucyl-L-leucyl dipeptide oxidase[J]. Biochemistry, 2010, 49(34): 7282-7296. |
| 46 | CHEN H N, ZHOU H B, SUN T, et al. Identification of holrhizins E-Q reveals the diversity of nonribosomal lipopeptides in Paraburkholderia rhizoxinica [J]. Journal of Natural Products, 2020, 83(2): 537-541. |
| 47 | CHEN H N, SUN T, BAI X P, et al. Genomics-driven activation of silent biosynthetic gene clusters in Burkholderia gladioli by screening recombineering system[J]. Molecules, 2021, 26: 700. |
| [1] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| [2] | SONG Yongxiang, ZHANG Xiufeng, LI Yanqin, XIAO Hua, YAN Yan. Resistance-gene directed discovery of bioactive natural products [J]. Synthetic Biology Journal, 2024, 5(3): 474-491. |
| [3] | LEI Ru, TAO Hui, LIU Tiangang. Deep genome mining boosts the discovery of microbial terpenoids [J]. Synthetic Biology Journal, 2024, 5(3): 507-526. |
| [4] | XI Mengyu, HU Yiling, GU Yucheng, GE Huiming. Genome mining-directed discovery for natural medicinal products [J]. Synthetic Biology Journal, 2024, 5(3): 447-473. |
| [5] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [6] | LIN Jicong, ZOU Gen, LIU Hongmin, WEI Yongjun. Application of CRISPR/Cas genome editing technology in the synthesis of secondary metabolites of filamentous fungi [J]. Synthetic Biology Journal, 2023, 4(4): 738-755. |
| [7] | ZHANG Fanzhong, XIANG Changjun, ZHANG Lihan. Advances and applications of evolutionary analysis and big-data guided bioinformatics in natural product research [J]. Synthetic Biology Journal, 2023, 4(4): 629-650. |
| [8] | LAI Mingyuan, WEI Jian, XU Jianhe, YU Huilei. Review of research on unspecific peroxygenases (UPOs) [J]. Synthetic Biology Journal, 2022, 3(6): 1235-1249. |
| [9] | DONG Jiayu, LI Min, XIAO Zonghua, HU Ming, MATSUDA Yudai, WANG Weiguang. Recent advances in heterologous production of natural products using Aspergillus oryzae [J]. Synthetic Biology Journal, 2022, 3(6): 1126-1149. |
| [10] | YANG Qian, CHENG Botao, TANG Zhijun, LIU Wen. Applications and prospects of genome mining in the discovery of natural products [J]. Synthetic Biology Journal, 2021, 2(5): 697-715. |
| [11] | Zhengjie HOU, Huizhong SUN, Song BAI, Xinyue CHEN, Chunyang CAO, Jingsheng CHENG. Research progress of cyclic lipopeptide biosynthesis [J]. Synthetic Biology Journal, 2021, 2(4): 577-597. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||