Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (3): 507-526.DOI: 10.12211/2096-8280.2023-098
• Invited Review • Previous Articles Next Articles
Deep genome mining boosts the discovery of microbial terpenoids
LEI Ru, TAO Hui, LIU Tiangang
- Key Laboratory of Combinatorial Biosynthesis and Drug Discovery,Ministry of Education,School of Pharmaceutical Sciences,Wuhan University,Wuhan 430071,Hubei,China
-
Received:2023-12-01Revised:2024-02-22Online:2024-07-12Published:2024-06-30 -
Contact:TAO Hui, LIU Tiangang
基因组深度挖掘驱动微生物萜类化合物高效发现
雷茹, 陶慧, 刘天罡
- 武汉大学药学院,组合生物合成与新药发现教育部重点实验室,湖北 武汉 430071
-
通讯作者:陶慧,刘天罡 -
作者简介:雷茹 (1998—),女,硕士研究生。研究方向为深海来源真菌及放线菌基因组测序与功能分析。E-mail:2017302290014@whu.edu.cn陶慧 (1990—),女,教授,博士生导师。研究方向为复杂微生物来源天然产物的生物合成机制解析与绿色生物制造。E-mail:thui@whu.edu.cn刘天罡 (1979—),男,教授,博士生导师。研究方向为萜类等天然产物的高效合成与创新发现;基于底盘细胞和自动化平台的天然产物基因组挖掘;微生物与人体的代谢互作。E-mail:liutg@whu.edu.cn -
基金资助:国家重点研发计划(2023YFA0916200)
CLC Number:
Cite this article
LEI Ru, TAO Hui, LIU Tiangang. Deep genome mining boosts the discovery of microbial terpenoids[J]. Synthetic Biology Journal, 2024, 5(3): 507-526.
雷茹, 陶慧, 刘天罡. 基因组深度挖掘驱动微生物萜类化合物高效发现[J]. 合成生物学, 2024, 5(3): 507-526.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-098
| 主要微生物底盘 | 优点 | 缺点 |
|---|---|---|
| Escherichia coli | 遗传操作简单 培养周期短 具有内源MEP途径 | 不具有跨膜结构域,可能导致功能蛋白的错误折叠、失去活性甚至降解 不适合表达真菌和植物来源的萜类合成基因(簇) |
| Saccharomyces cerevisiae | 遗传操作相对简易 重组效率高效 具有完整的细胞器与内膜系统 具有内源MVA途径 | 存在密码子偏好性 无法正确识别和剪切内含子 不适合表达真菌来源的萜类合成基因(簇) |
| Aspergillus oryzae | 具有强大的蛋白分泌特性 功能酶后修饰能力强大 可以正确地识别和剪切内含子 具有内源MVA途径 适合表达真菌或植物来源的萜类合成基因(簇) | 遗传操作复杂 培养周期长 |
| Streptomyces albus | 功能酶后修饰能力强大 具有内源MEP途径 适合表达细菌来源的萜类合成基因(簇) | 遗传操作复杂 培养周期长 |
Table 1 Major microbial chassis for terpenoid mining
| 主要微生物底盘 | 优点 | 缺点 |
|---|---|---|
| Escherichia coli | 遗传操作简单 培养周期短 具有内源MEP途径 | 不具有跨膜结构域,可能导致功能蛋白的错误折叠、失去活性甚至降解 不适合表达真菌和植物来源的萜类合成基因(簇) |
| Saccharomyces cerevisiae | 遗传操作相对简易 重组效率高效 具有完整的细胞器与内膜系统 具有内源MVA途径 | 存在密码子偏好性 无法正确识别和剪切内含子 不适合表达真菌来源的萜类合成基因(簇) |
| Aspergillus oryzae | 具有强大的蛋白分泌特性 功能酶后修饰能力强大 可以正确地识别和剪切内含子 具有内源MVA途径 适合表达真菌或植物来源的萜类合成基因(簇) | 遗传操作复杂 培养周期长 |
| Streptomyces albus | 功能酶后修饰能力强大 具有内源MEP途径 适合表达细菌来源的萜类合成基因(簇) | 遗传操作复杂 培养周期长 |
| 数据库或网络工具 | 简介 | 网址 | 参考文献 |
|---|---|---|---|
| BGC数据库 | |||
| antiSMASH Database | 细菌、古细菌和真菌等基因组中可检测的高质量BGC集合 | https://antismash-db.secondarymetabolites.org/ | [ |
| BiG-FAM | 微生物生物合成基因簇家族(GCF)模型集合 | https://bigfam.bioinformatics.nl/home | [ |
| IMG-ABC | 微生物BGC的集成基因组图谱 | https://img.jgi.doe.gov/cgi-bin/abc-public/main.cgi | [ |
| MIBiG | 已知生物合成基因簇及其分子产物的信息集合 | https://mibig.secondarymetabolites.org/ | [ |
| BGC网络识别工具 | |||
| antiSMASH | 用于自动识别和分析微生物次级代谢产物基因簇 | https://antismash.secondarymetabolites.org/ | [ |
| ARTS | 细菌基因组靶向挖掘,筛选潜在新型抗生素靶点 | https://arts.ziemertlab.com/ | [ |
| BiG-SCAPE | 用于构建BGC的序列相似性网络并将其分组到GCF中 | https://git.wageningenur.nl/medema-group/BiG-SCAPE/wikis/home | [ |
| BiG-SLiCE | 聚集大量BGC,通过序列相似性网络构建GCF模型 | https://github.com/medema-group/bigslice | [ |
| ClusterFinder | 预测基因组中的BGC | https://github.com/petercim/ClusterFinder | [ |
| DeepBGC | 使用深度学习的方法预测细菌和真菌基因组中的BGC | https://github.com/Merck/deepbgc | [ |
| e-DeepBGC | 基于深度学习方法,引入了Pfam信息,预测细菌基因组中BGC | — | [ |
| RL-BGC | 基于Pfam蛋白质家族结构域和功能注释的强化学习方法,精准获得真菌候选BGC | https://github.com/bioinfoUQAM/RL-bgc-components | [ |
| PRISM4 | 基于微生物基因组序列,识别BGC并生成结构预测的组合文库 | https://prism.adapsyn.com/ | [ |
| SMURF | 用于真菌基因组BGC和途径预测分析 | http://smurf.jcvi.org/index.php | [ |
| TeroKit | 萜类化合物的化学空间、生物活性和生物合成途径的在线检索和分析 | http://terokit.qmclab.com/ | [ |
| 酶功能预测工具 | |||
| CLEAN | 启用对比学习的酶注释,能够对未表征的酶实现功能预测 | https://clean.platform.moleculemaker.org/configuration | [ |
| ECRECer | 基于深度学习实现酶催化功能预测 | https://ecrecer.biodesign.ac.cn/ | [ |
| PfamScan | 根据Pfam HMM数据库进行酶功能预测分析 | https://www.ebi.ac.uk/Tools/pfa/pfamscan/ | [ |
Table 2 Tools and databases for mining terpene BGCs [104-106]
| 数据库或网络工具 | 简介 | 网址 | 参考文献 |
|---|---|---|---|
| BGC数据库 | |||
| antiSMASH Database | 细菌、古细菌和真菌等基因组中可检测的高质量BGC集合 | https://antismash-db.secondarymetabolites.org/ | [ |
| BiG-FAM | 微生物生物合成基因簇家族(GCF)模型集合 | https://bigfam.bioinformatics.nl/home | [ |
| IMG-ABC | 微生物BGC的集成基因组图谱 | https://img.jgi.doe.gov/cgi-bin/abc-public/main.cgi | [ |
| MIBiG | 已知生物合成基因簇及其分子产物的信息集合 | https://mibig.secondarymetabolites.org/ | [ |
| BGC网络识别工具 | |||
| antiSMASH | 用于自动识别和分析微生物次级代谢产物基因簇 | https://antismash.secondarymetabolites.org/ | [ |
| ARTS | 细菌基因组靶向挖掘,筛选潜在新型抗生素靶点 | https://arts.ziemertlab.com/ | [ |
| BiG-SCAPE | 用于构建BGC的序列相似性网络并将其分组到GCF中 | https://git.wageningenur.nl/medema-group/BiG-SCAPE/wikis/home | [ |
| BiG-SLiCE | 聚集大量BGC,通过序列相似性网络构建GCF模型 | https://github.com/medema-group/bigslice | [ |
| ClusterFinder | 预测基因组中的BGC | https://github.com/petercim/ClusterFinder | [ |
| DeepBGC | 使用深度学习的方法预测细菌和真菌基因组中的BGC | https://github.com/Merck/deepbgc | [ |
| e-DeepBGC | 基于深度学习方法,引入了Pfam信息,预测细菌基因组中BGC | — | [ |
| RL-BGC | 基于Pfam蛋白质家族结构域和功能注释的强化学习方法,精准获得真菌候选BGC | https://github.com/bioinfoUQAM/RL-bgc-components | [ |
| PRISM4 | 基于微生物基因组序列,识别BGC并生成结构预测的组合文库 | https://prism.adapsyn.com/ | [ |
| SMURF | 用于真菌基因组BGC和途径预测分析 | http://smurf.jcvi.org/index.php | [ |
| TeroKit | 萜类化合物的化学空间、生物活性和生物合成途径的在线检索和分析 | http://terokit.qmclab.com/ | [ |
| 酶功能预测工具 | |||
| CLEAN | 启用对比学习的酶注释,能够对未表征的酶实现功能预测 | https://clean.platform.moleculemaker.org/configuration | [ |
| ECRECer | 基于深度学习实现酶催化功能预测 | https://ecrecer.biodesign.ac.cn/ | [ |
| PfamScan | 根据Pfam HMM数据库进行酶功能预测分析 | https://www.ebi.ac.uk/Tools/pfa/pfamscan/ | [ |
| 71 | TANG M C, SHEN C, DENG Z X, et al. Combinatorial biosynthesis of terpenoids through mixing-and-matching sesquiterpene cyclase and cytochrome P450 pairs[J]. Organic Letters, 2022, 24(26): 4783-4787. |
| 72 | TSUKADA K, SHINKI S, KANEKO A, et al. Synthetic biology based construction of biological activity-related library of fungal decalin-containing diterpenoid pyrones[J]. Nature Communications, 2020, 11(1): 1830. |
| 73 | DUAN Y T, KOUTSAVITI A, HARIZANI M, et al. Widespread biosynthesis of 16-carbon terpenoids in bacteria[J]. Nature Chemical Biology, 2023, 19(12): 1532-1539. |
| 74 | ROTHSCHILD L J, MANCINELLI R L. Life in extreme environments[J]. Nature, 2001, 409: 1092-1101. |
| 75 | SHU W S, HUANG L N. Microbial diversity in extreme environments[J]. Nature Reviews Microbiology, 2022, 20(4): 219-235. |
| 76 | GARETH JONES E B, PANG K L, ABDEL-WAHAB M A, et al. An online resource for marine fungi[J]. Fungal Diversity, 2019, 96(1): 347-433. |
| 77 | GONÇALVES M F M, ESTEVES A C, ALVES A. Marine fungi: opportunities and challenges[J]. Encyclopedia, 2022, 2(1): 559-577. |
| 78 | SHABANA S, LAKSHMI K R, SATYA A K. An updated review of secondary metabolites from marine fungi[J]. Mini Reviews in Medicinal Chemistry, 2021, 21(5): 602-642. |
| 79 | JIANG M H, WU Z E, GUO H, et al. A review of terpenes from marine-derived fungi: 2015-2019[J]. Marine Drugs, 2020, 18(6): 321. |
| 80 | YANG X W, PENG K, LIU Z, et al. Strepsesquitriol, a rearranged zizaane-type sesquiterpenoid from the deep-sea-derived actinomycete Streptomyces sp. SCSIO 10355[J]. Journal of Natural Products, 2013, 76(12): 2360-2363. |
| 81 | KIM D, LEE E J, LEE J, et al. Antartin, a cytotoxic zizaane-type sesquiterpenoid from a Streptomyces sp. isolated from an Antarctic marine sediment[J]. Marine Drugs, 2018, 16(4): 130. |
| 82 | CARROLL A R, COPP B R, DAVIS R A, et al. Marine natural products[J]. Natural Product Reports, 2023,40(2):275-325. |
| 83 | CARROLL A R, COPP B R, DAVIS R A, et al. Marine natural products[J]. Natural Product Reports, 2022,39(6):1122-1171. |
| 84 | CARROLL A R, COPP B R, DAVIS R A, et al. Marine natural products[J]. Natural Product Reports, 2021,38(2):362-413. |
| 85 | SCHULTZ J, MODOLON F, PEIXOTO R S, et al. Shedding light on the composition of extreme microbial dark matter: alternative approaches for culturing extremophiles[J]. Frontiers in Microbiology, 2023, 14: 1167718. |
| 86 | GUO J J, CAI Y S, CHENG F C, et al. Genome mining reveals a multiproduct sesterterpenoid biosynthetic gene cluster in Aspergillus ustus [J]. Organic Letters, 2021, 23(5): 1525-1529. |
| 87 | ZHANG P, WU G W, HEARD S C, et al. Identification and characterization of a cryptic bifunctional type Ⅰ diterpene synthase involved in talaronoid biosynthesis from a marine-derived fungus[J]. Organic Letters, 2022, 24(38): 7037-7041. |
| 88 | 曲戈, 朱彤, 蒋迎迎, 等. 蛋白质工程:从定向进化到计算设计[J]. 生物工程学报, 2019, 35(10): 1843-1856. |
| QU G, ZHU T, JIANG Y Y, et al. Protein engineering: from directed evolution to computational design[J]. Chinese Journal of Biotechnology, 2019, 35(10): 1843-1856. | |
| 89 | 卞佳豪, 杨广宇. 人工智能辅助的蛋白质工程[J]. 合成生物学, 2022, 3(3): 429-444. |
| BIAN J H, YANG G Y. Artificial intelligence-assisted protein engineering[J]. Synthetic Biology Journal, 2022, 3(3): 429-444. | |
| 90 | LI Z, ZHANG L L, XU K W, et al. Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase[J]. Nature Communications, 2023, 14(1): 4001. |
| 91 | IGNEA C, PONTINI M, MOTAWIA M S, et al. Synthesis of 11-carbon terpenoids in yeast using protein and metabolic engineering[J]. Nature Chemical Biology, 2018, 14(12): 1090-1098. |
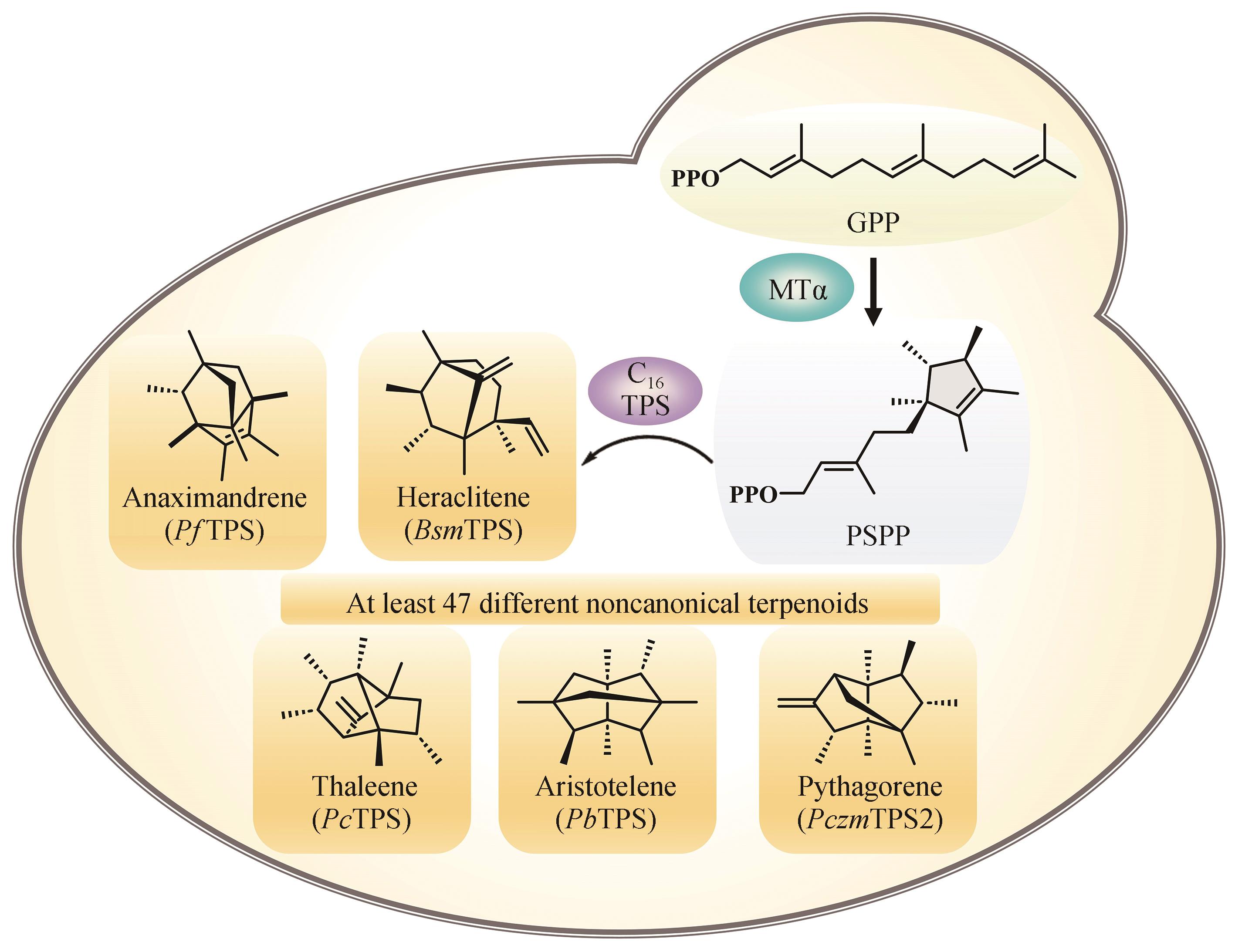
| 92 | IGNEA C, RAADAM M H, KOUTSAVITI A, et al. Expanding the terpene biosynthetic code with non-canonical 16 carbon atom building blocks[J]. Nature Communications, 2022, 13(1): 5188. |
| 93 | HAMEDIRAD M, CHAO R, WEISBERG S, et al. Towards a fully automated algorithm driven platform for biosystems design[J]. Nature Communications, 2019, 10(1): 5150. |
| 94 | ENGHIAD B, XUE P, SINGH N, et al. PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction[J]. Nature Communications, 2022, 13(1): 2697. |
| 95 | LEFERINK N G H, DUNSTAN M S, HOLLYWOOD K A, et al. An automated pipeline for the screening of diverse monoterpene synthase libraries[J]. Scientific Reports, 2019, 9(1): 11936. |
| 96 | YUAN W, JIANG C J, WANG Q, et al. Biosynthesis of mushroom-derived type Ⅱ ganoderic acids by engineered yeast[J]. Nature Communications, 2022, 13(1): 7740. |
| 97 | 崔金明, 张炳照, 马迎飞, 等. 合成生物学研究的工程化平台[J]. 中国科学院院刊, 2018, 33(11): 1249-1257. |
| CUI J M, ZHANG B Z, MA Y F, et al. Engineering platforms for synthetic biology research[J]. Bulletin of Chinese Academy of Sciences, 2018, 33(11): 1249-1257. | |
| 98 | 卢挥, 张芳丽, 黄磊. 合成生物学自动化装置iBioFoundry的构建与应用[J]. 合成生物学, 2023, 4(5): 877-891. |
| LU H, ZHANG F L, HUANG L. Establishment of iBioFoundry for synthetic biology applications[J]. Synthetic Biology Journal, 2023, 4(5): 877-891. | |
| 99 | 张亭, 冷梦甜, 金帆, 等. 合成生物研究重大科技基础设施概述[J]. 合成生物学, 2022, 3(1): 184-194. |
| ZHANG T, LENG M T, JIN F, et al. Overview on platform for synthetic biology research at Shenzhen[J]. Synthetic Biology Journal, 2022, 3(1): 184-194. | |
| 100 | SCHERLACH K, HERTWECK C. Mining and unearthing hidden biosynthetic potential[J]. Nature Communications, 2021, 12(1): 3864. |
| 101 | HEMMERLING F, PIEL J. Strategies to access biosynthetic novelty in bacterial genomes for drug discovery[J]. Nature Reviews Drug Discovery, 2022, 21(5): 359-378. |
| 102 | ZENG T, CHEN Y X X, JIAN Y X, et al. Chemotaxonomic investigation of plant terpenoids with an established database (TeroMOL)[J]. The New Phytologist, 2022, 235(2): 662-673. |
| 1 | ZENG T, LIU Z H, ZHUANG J Y, et al. TeroKit: a database-driven web server for terpenome research[J]. Journal of Chemical Information and Modeling, 2020, 60(4): 2082-2090. |
| 2 | FORDJOUR E, MENSAH E O, HAO Y P, et al. Toward improved terpenoids biosynthesis: strategies to enhance the capabilities of cell factories[J]. Bioresources and Bioprocessing, 2022, 9(1): 6. |
| 3 | AVALOS M, GARBEVA P, VADER L, et al. Biosynthesis, evolution and ecology of microbial terpenoids[J]. Natural Product Reports, 2022, 39(2): 249-272. |
| 4 | ARNESEN J A, JACOBSEN I H, DYEKJÆR J D, et al. Production of abscisic acid in the oleaginous yeast Yarrowia lipolytica [J]. FEMS Yeast Research, 2022, 22(1): foac015. |
| 5 | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 6 | WU Q Y, HUANG Z Y, WANG J Y, et al. Construction of an Escherichia coli cell factory to synthesize taxadien-5α-ol, the key precursor of anti-cancer drug paclitaxel[J]. Bioresources and Bioprocessing, 2022, 9(1): 82. |
| 7 | ALBERTI F, KHAIRUDIN K, DAVIES J A, et al. Biosynthesis of pleuromutilin congeners using an Aspergillus oryzae expression platform[J]. Chemical Science, 2023, 14(14): 3826-3833. |
| 8 | HOU A W, LAUTERBACH L, DICKSCHAT J S. Enzymatic synthesis of methylated terpene analogues using the plasticity of bacterial terpene synthases[J]. Chemistry, 2020, 26(10): 2178-2182. |
| 9 | HOU A W, DICKSCHAT J S. Using terpene synthase plasticity in catalysis: on the enzymatic conversion of synthetic farnesyl diphosphate analogues[J]. Chemistry, 2021, 27(63): 15644-15649. |
| 10 | VON REUSS S H, KAI M, PIECHULLA B, et al. Octamethylbicyclo[3.2.1]octadienes from the rhizobacterium Serratia odorifera [J]. Angewandte Chemie International Edition, 2010, 49(11): 2009-2010. |
| 11 | KOMATSU M, TSUDA M, OMURA S, et al. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(21): 7422-7427. |
| 12 | WANG C M, CANE D E. Biochemistry and molecular genetics of the biosynthesis of the earthy odorant methylisoborneol in Streptomyces coelicolor [J]. Journal of the American Chemical Society, 2008, 130(28): 8908-8909. |
| 13 | VON REUSS S, DOMIK D, LEMFACK M C, et al. Sodorifen biosynthesis in the rhizobacterium Serratia plymuthica involves methylation and cyclization of MEP-derived farnesyl pyrophosphate by a SAM-dependent C-methyltransferase[J]. Journal of the American Chemical Society, 2018, 140(37): 11855-11862. |
| 103 | CHEN N H, ZHANG R, ZENG T, et al. Developing TeroENZ and TeroMAP modules for the terpenome research platform TeroKit[J]. Database, 2023, 2023: baad020. |
| 104 | 赖奇龙, 姚帅, 查毓国, 等. 微生物组生物合成基因簇发掘方法及应用前景[J]. 合成生物学, 2023, 4(3): 611-627. |
| LAI Q L, YAO S, ZHA Y G, et al. Microbiome-based biosynthetic gene cluster data mining techniques and application potentials[J]. Synthetic Biology Journal, 2023, 4(3): 611-627. | |
| 105 | 杨谦, 程伯涛, 汤志军, 等. 基因组挖掘在天然产物发现中的应用和前景[J]. 合成生物学, 2021, 2(5): 697-715. |
| YANG Q, CHENG B T, TANG Z J, et al. Applications and prospects of genome mining in the discovery of natural products[J]. Synthetic Biology Journal, 2021, 2(5): 697-715. | |
| 106 | REN H Q, SHI C Y, ZHAO H M. Computational tools for discovering and engineering natural product biosynthetic pathways[J]. iScience, 2020, 23(1): 100795. |
| 107 | BLIN K, SHAW S, MEDEMA M H, et al. The antiSMASH database version 4: additional genomes and BGCs, new sequence-based searches and more[J]. Nucleic Acids Research, 2024, 52(D1): D586-D589. |
| 108 | KAUTSAR S A, BLIN K, SHAW S, et al. BiG-FAM: the biosynthetic gene cluster families database[J]. Nucleic Acids Research, 2021, 49(D1): D490-D497. |
| 109 | PALANIAPPAN K, CHEN I M A, CHU K, et al. IMG-ABC v.5.0: an update to the IMG/Atlas of Biosynthetic Gene Clusters Knowledgebase[J]. Nucleic Acids Research, 2020, 48(D1): D422-D430. |
| 110 | TERLOUW B R, BLIN K, NAVARRO-MUÑOZ J C, et al. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters[J]. Nucleic Acids Research, 2023, 51(D1): D603-D610. |
| 111 | BLIN K, SHAW S, AUGUSTIJN H E, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation[J]. Nucleic Acids Research, 2023, 51(W1): W46-W50. |
| 112 | MUNGAN M D, ALANJARY M, BLIN K, et al. ARTS 2.0: feature updates and expansion of the Antibiotic Resistant Target Seeker for comparative genome mining[J]. Nucleic Acids Research, 2020, 48(W1): W546-W552. |
| 113 | NAVARRO-MUÑOZ J C, SELEM-MOJICA N, MULLOWNEY M W, et al. A computational framework to explore large-scale biosynthetic diversity[J]. Nature Chemical Biology, 2020, 16(1): 60-68. |
| 114 | KAUTSAR S A, VAN DER HOOFT J J J, DE RIDDER D, et al. BiG-SLiCE: a highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters[J]. GigaScience, 2021, 10(1): giaa154. |
| 115 | CIMERMANCIC P, MEDEMA M H, CLAESEN J, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters[J]. Cell, 2014, 158(2): 412-421. |
| 116 | HANNIGAN G D, PRIHODA D, PALICKA A, et al. A deep learning genome-mining strategy for biosynthetic gene cluster prediction[J]. Nucleic Acids Research, 2019, 47(18): e110. |
| 117 | LIU M Y, LI Y, LI H Z. Deep learning to predict the biosynthetic gene clusters in bacterial genomes[J]. Journal of Molecular Biology, 2022, 434(15): 167597. |
| 118 | ALMEIDA H, TSANG A, DIALLO A B. Improving candidate Biosynthetic Gene Clusters in fungi through reinforcement learning[J]. Bioinformatics, 2022, 38(16): 3984-3991. |
| 119 | SKINNIDER M A, JOHNSTON C W, GUNABALASINGAM M, et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences[J]. Nature Communications, 2020, 11(1): 6058. |
| 120 | KHALDI N, SEIFUDDIN F T, TURNER G, et al. SMURF: genomic mapping of fungal secondary metabolite clusters[J]. Fungal Genetics and Biology, 2010, 47(9): 736-741. |
| 121 | YU T H, CUI H Y, LI J C, et al. Enzyme function prediction using contrastive learning[J]. Science, 2023, 379(6639): 1358-1363. |
| 122 | SHI Z K, DENG R, YUAN Q Q, et al. Enzyme commission number prediction and benchmarking with hierarchical dual-core multitask learning framework[J]. Research, 2023, 6: 153. |
| 123 | LI W Z, COWLEY A, ULUDAG M, et al. The EMBL-EBI bioinformatics web and programmatic tools framework[J]. Nucleic Acids Research, 2015, 43(W1): W580-W584. |
| 124 | MISTRY J, BATEMAN A, FINN R D. Predicting active site residue annotations in the Pfam database[J]. BMC Bioinformatics, 2007, 8: 298. |
| 125 | SAHAYASHEELA V J, LANKADASARI M B, DAN V M, et al. Artificial intelligence in microbial natural product drug discovery: current and emerging role[J]. Natural Product Reports, 2022, 39(12): 2215-2230. |
| 14 | HU Z X, YE Y, ZHANG Y H. Large-scale culture as a complementary and practical method for discovering natural products with novel skeletons[J]. Natural Product Reports, 2021, 38(10): 1775-1793. |
| 15 | RYU M J, HWANG S, KIM S, et al. Meroindenon and merochlorins E and F, antibacterial meroterpenoids from a marine-derived sediment bacterium of the genus Streptomyces [J]. Organic Letters, 2019, 21(15): 5779-5783. |
| 16 | ZHAI G F, CHEN S H, SHEN H J, et al. Bioactive monoterpenes and polyketides from the ascidian-derived fungus Diaporthe sp. SYSU-MS4722[J]. Marine Drugs, 2022, 20(9): 553. |
| 17 | LIU H, PU Y H, REN J W, et al. Genetic dereplication driven discovery of a tricinoloniol acid biosynthetic pathway in Trichoderma hypoxylon [J]. Organic & Biomolecular Chemistry, 2020, 18(28): 5344-5348. |
| 18 | KIM S H, LU W L, AHMADI M K, et al. Atolypenes, tricyclic bacterial sesterterpenes discovered using a multiplexed in vitro Cas9-TAR gene cluster refactoring approach[J]. ACS Synthetic Biology, 2019, 8(1): 109-118. |
| 19 | SHEN X T, MO X H, ZHU L P, et al. Unusual and highly bioactive sesterterpenes synthesized by Pleurotus ostreatus during coculture with Trametes robiniophila Murr[J]. Applied and Environmental Microbiology, 2019, 85(14): e00293-19. |
| 20 | CHIANG C Y, OHASHI M, TANG Y. Deciphering chemical logic of fungal natural product biosynthesis through heterologous expression and genome mining[J]. Natural Product Reports, 2023, 40(1): 89-127. |
| 21 | MENG X F, FANG Y, DING M Y, et al. Developing fungal heterologous expression platforms to explore and improve the production of natural products from fungal biodiversity[J]. Biotechnology Advances, 2022, 54: 107866. |
| 22 | MATSUDA Y, MITSUHASHI T, LEE S K, et al. Astellifadiene: structure determination by NMR spectroscopy and crystalline sponge method, and elucidation of its biosynthesis[J]. Angewandte Chemie International Edition, 2016, 55(19): 5785-5788. |
| 23 | 程术, 邓子新, 卞光凯, 等. 萜类高效合成平台的搭建与萜类产物批量挖掘[J]. 生命科学, 2019, 31(5): 449-457. |
| CHENG S, DENG Z X, BIAN G K, et al. Construction of high-efficient terpenoid platform and the application in terpenoid discovery[J]. Chinese Bulletin of Life Sciences, 2019, 31(5): 449-457. | |
| 24 | ZHU F Y, ZHONG X F, HU M Z, et al. In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli [J]. Biotechnology and Bioengineering, 2014, 111(7): 1396-1405. |
| 25 | ZHU F Y, LU L, FU S, et al. Targeted engineering and scale up of lycopene overproduction in Escherichia coli [J]. Process Biochemistry, 2015, 50(3): 341-346. |
| 126 | MULLOWNEY M W, DUNCAN K R, ELSAYED S S, et al. Artificial intelligence for natural product drug discovery[J]. Nature Reviews Drug Discovery, 2023, 22(11): 895-916. |
| 127 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 128 | SASAKI D, FUJIHASHI M, OKUYAMA N, et al. Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation[J]. The Journal of Biological Chemistry, 2011, 286(5): 3729-3740. |
| 129 | TAO H, LAUTERBACH L, BIAN G K, et al. Discovery of non-squalene triterpenes[J]. Nature, 2022, 606(7913): 414-419. |
| 130 | KANG W, MA T, LIU M, et al. Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux[J]. Nature Communications, 2019, 10(1): 4248. |
| 131 | SUN X X, YUAN Y J, CHEN Q T, et al. Metabolic pathway assembly using docking domains from type Ⅰ cis-AT polyketide synthases[J]. Nature Communications, 2022, 13(1): 5541. |
| 132 | TIAN P F, WANG J, SHEN X L, et al. Fundamental CRISPR-Cas9 tools and current applications in microbial systems[J]. Synthetic and Systems Biotechnology, 2017, 2(3): 219-225. |
| 133 | LI Z H, LIU M, LYU X M, et al. CRISPR/Cpf1 facilitated large fragment deletion in Saccharomyces cerevisiae [J]. Journal of Basic Microbiology, 2018, 58(12): 1100-1104. |
| 134 | LI L, WEI K K, ZHENG G S, et al. CRISPR-Cpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces [J]. Applied and Environmental Microbiology, 2018, 84(18): e00827-18. |
| 135 | WU S M, TIAN P F, TAN T W. CRISPR-Cas13 technology portfolio and alliance with other genetic tools[J]. Biotechnology Advances, 2022, 61: 108047. |
| 136 | YANG Y, MAO Y F, WANG R Y, et al. AutoESD: a web tool for automatic editing sequence design for genetic manipulation of microorganisms[J]. Nucleic Acids Research, 2022, 50(W1): W75-W82. |
| 137 | CAESAR L K, MONTASER R, KELLER N P, et al. Metabolomics and genomics in natural products research: complementary tools for targeting new chemical entities[J]. Natural Product Reports, 2021, 38(11): 2041-2065. |
| 26 | MA T, ZHOU Y J, LI X W, et al. Genome mining of astaxanthin biosynthetic genes from Sphingomonas sp. ATCC 55669 for heterologous overproduction in Escherichia coli [J]. Biotechnology Journal, 2016, 11(2): 228-237. |
| 27 | BIAN G K, YUAN Y J, TAO H, et al. Production of taxadiene by engineering of mevalonate pathway in Escherichia coli and endophytic fungus Alternaria alternata TPF6[J]. Biotechnology Journal, 2017, 12(4): 1600697. |
| 28 | BIAN G K, DENG Z X, LIU T G. Strategies for terpenoid overproduction and new terpenoid discovery[J]. Current Opinion in Biotechnology, 2017, 48: 234-241. |
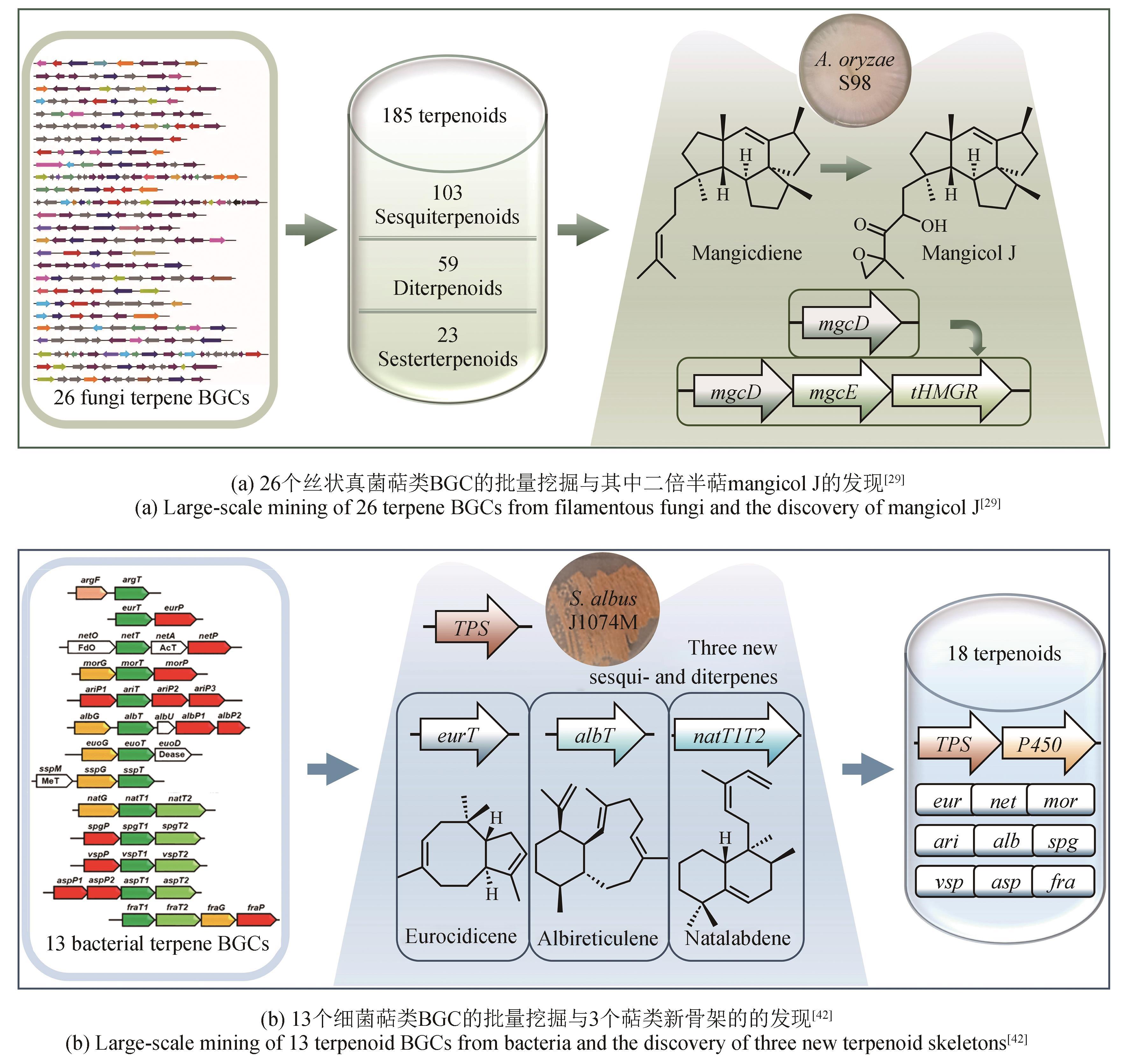
| 29 | YUAN Y J, CHENG S, BIAN G K, et al. Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi[J]. Nature Catalysis, 2022, 5: 277-287. |
| 30 | BIAN G K, HAN Y C, HOU A W, et al. Releasing the potential power of terpene synthases by a robust precursor supply platform[J]. Metabolic Engineering, 2017, 42: 1-8. |
| 31 | BIAN G K, HOU A W, YUAN Y J, et al. Metabolic engineering-based rapid characterization of a sesquiterpene cyclase and the skeletons of fusariumdiene and fusagramineol from Fusarium graminearum [J]. Organic Letters, 2018, 20(6): 1626-1629. |
| 32 | BIAN G K, RINKEL J, WANG Z Q, et al. A clade Ⅱ-D fungal chimeric diterpene synthase from Colletotrichum gloeosporioides produces dolasta-1(15),8-diene[J]. Angewandte Chemie International Edition, 2018, 57(48): 15887-15890. |
| 33 | SIEMON T, WANG Z Q, BIAN G K, et al. Semisynthesis of plant-derived englerin A enabled by microbe engineering of guaia-6,10(14)-diene as building block[J]. Journal of the American Chemical Society, 2020, 142(6): 2760-2765. |
| 34 | ZUO Y M, XIAO F, GAO J C, et al. Establishing Komagataella phaffii as a cell factory for efficient production of sesquiterpenoid α-santalene[J]. Journal of Agricultural and Food Chemistry, 2022, 70(26): 8024-8031. |
| 35 | LUO G J, LIN Y, CHEN S T, et al. Overproduction of patchoulol in metabolically engineered Komagataella phaffii [J]. Journal of Agricultural and Food Chemistry, 2023, 71(4): 2049-2058. |
| 36 | LI Z J, WANG Y Z, WANG L R, et al. Advanced strategies for the synthesis of terpenoids in Yarrowia lipolytica [J]. Journal of Agricultural and Food Chemistry, 2021, 69(8): 2367-2381. |
| 37 | MA Y R, WANG K F, WANG W J, et al. Advances in the metabolic engineering of Yarrowia lipolytica for the production of terpenoids[J]. Bioresource Technology, 2019, 281: 449-456. |
| 38 | ZHANG T L, YU H W, YE L D. Metabolic engineering of Yarrowia lipolytica for terpenoid production: tools and strategies[J]. ACS Synthetic Biology, 2023, 12(3): 639-656. |
| 39 | ZHANG G, WANG H, ZHANG Z, et al. Metabolic engineering of Yarrowia lipolytica for terpenoids production: advances and perspectives[J]. Critical Reviews in Biotechnology, 2022, 42(4): 618-633. |
| 40 | LIN S Y, OAKLEY C E, JENKINSON C B, et al. A heterologous expression platform in Aspergillus nidulans for the elucidation of cryptic secondary metabolism biosynthetic gene clusters: discovery of the Aspergillus fumigatus sartorypyrone biosynthetic pathway[J]. Chemical Science, 2023, 14(40): 11022-11032. |
| 41 | TARASOVA E V, LUCHNIKOVA N A, GRISHKO V V, et al. Actinomycetes as producers of biologically active terpenoids: current trends and patents[J]. Pharmaceuticals, 2023, 16(6): 872. |
| 42 | HU Y L, ZHANG Q, LIU S H, et al. Building Streptomyces albus as a chassis for synthesis of bacterial terpenoids[J]. Chemical Science, 2023, 14(13): 3661-3667. |
| 43 | MYRONOVSKYI M, LUZHETSKYY A. Heterologous production of small molecules in the optimized Streptomyces hosts[J]. Natural Product Reports, 2019, 36(9): 1281-1294. |
| 44 | AJIKUMAR P K, XIAO W H, TYO K E, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli [J]. Science, 2010, 330(6000): 70-74. |
| 45 | REDDY G K, LEFERINK N G H, UMEMURA M, et al. Exploring novel bacterial terpene synthases[J]. PLoS One, 2020, 15(4): e0232220. |
| 46 | LIN G M, VOIGT C A. Design of a redox-proficient Escherichia coli for screening terpenoids and modifying cytochrome P450s[J]. Nature Catalysis, 2023, 6: 1016-1029. |
| 47 | NAVALE G R, DHARNE M S, SHINDE S S. Metabolic engineering and synthetic biology for isoprenoid production in Escherichia coli and Saccharomyces cerevisiae [J]. Applied Microbiology and Biotechnology, 2021, 105(2): 457-475. |
| 48 | CHEN R, JIA Q D, MU X, et al. Systematic mining of fungal chimeric terpene synthases using an efficient precursor-providing yeast chassis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(29): e2023247118. |
| 49 | 李晓东, 杨成帅, 王平平, 等. 构建酿酒酵母细胞工厂从头合成倍半萜类化合物α-新丁香三环烯和β-石竹烯[J]. 合成生物学, 2021, 2(5): 792-803. |
| 138 | DUNCAN K R, CRÜSEMANN M, LECHNER A, et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species[J]. Chemistry & Biology, 2015, 22(4): 460-471. |
| 139 | MAANSSON M, VYNNE N G, KLITGAARD A, et al. An integrated metabolomic and genomic mining workflow to uncover the biosynthetic potential of bacteria[J]. mSystems, 2016, 1(3): e00028-15. |
| 140 | TRYON J H, ROTE J C, CHEN L, et al. Genome mining and metabolomics uncover a rare D-capreomycidine containing natural product and its biosynthetic gene cluster[J]. ACS Chemical Biology, 2020, 15(11): 3013-3020. |
| 141 | LI C S, HU Y F, WU X H, et al. Discovery of unusual dimeric piperazyl cyclopeptides encoded by a Lentzea flaviverrucosa DSM 44664 biosynthetic supercluster[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(17): e2117941119. |
| 142 | CAESAR L K, BUTUN F A, ROBEY M T, et al. Correlative metabologenomics of 110 fungi reveals metabolite-gene cluster pairs[J]. Nature Chemical Biology, 2023, 19(7): 846-854. |
| 143 | RUDOLF J D, ALSUP T A, XU B F, et al. Bacterial terpenome[J]. Natural Product Reports, 2021, 38(5): 905-980. |
| 144 | IMHOFF J F. Natural products from marine fungi: still an underrepresented resource[J]. Marine Drugs, 2016, 14(1): 19. |
| 145 | YANG Z J, HE J Q, WEI X, et al. Exploration and genome mining of natural products from marine Streptomyces [J]. Applied Microbiology and Biotechnology, 2020, 104(1): 67-76. |
| 49 | LI X D, YANG C S, WANG P P, et al. Production of sesquiterpenoids α-neoclovene and β-caryophyllene by engineered Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2021, 2(5): 792-803. |
| 50 | DONG H, CHEN S, ZHU J X, et al. Enhance production of diterpenoids in yeast by overexpression of the fused enzyme of ERG20 and its mutant mERG20[J]. Journal of Biotechnology, 2020, 307: 29-34. |
| 51 | MUKHERJEE M, BLAIR R H, WANG Z Q. Machine-learning guided elucidation of contribution of individual steps in the mevalonate pathway and construction of a yeast platform strain for terpenoid production[J]. Metabolic Engineering, 2022, 74: 139-149. |
| 52 | MA Y S, ZU Y X, HUANG S W, et al. Engineering a universal and efficient platform for terpenoid synthesis in yeast[J]. Proceedings of the National Academy of Sciences of the United States of America, 2023, 120(1): e2207680120. |
| 53 | MEYER V, FIEDLER M, NITSCHE B, et al. The cell factory Aspergillus enters the big data era: opportunities and challenges for optimising product formation[J]. Advances in Biochemical Engineering/Biotechnology, 2015, 149: 91-132. |
| 54 | TONG Z Y, ZHENG X M, TONG Y, et al. Systems metabolic engineering for citric acid production by Aspergillus niger in the post-genomic era[J]. Microbial Cell Factories, 2019, 18(1): 28. |
| 55 | HUANG X N, CHEN M, LU X F, et al. Direct production of itaconic acid from liquefied corn starch by genetically engineered Aspergillus terreus [J]. Microbial Cell Factories, 2014, 13: 108. |
| 56 | HUANG X N, TANG S, ZHENG L H, et al. Construction of an efficient and robust Aspergillus terreus cell factory for monacolin J production[J]. ACS Synthetic Biology, 2019, 8(4): 818-825. |
| 57 | ICHISHIMA E. Development of enzyme technology for Aspergillus oryzae, A. sojae, and A. luchuensis, the national microorganisms of Japan[J]. Bioscience, Biotechnology, and Biochemistry, 2016, 80(9): 1681-1692. |
| 58 | VAN BERGEIJK D A, TERLOUW B R, MEDEMA M H, et al. Ecology and genomics of Actinobacteria: new concepts for natural product discovery[J]. Nature Reviews Microbiology, 2020, 18(10): 546-558. |
| 59 | 董雷, 韩嘉瑞, 李帅, 等. 链霉菌最新研究进展[J]. 微生物学报, 2023, 63(5): 1815-1832. |
| DONG L, HAN J R, LI S, et al. The latest research progress of streptomycetes[J]. Acta Microbiologica Sinica, 2023, 63(5): 1815-1832. | |
| 60 | AHMED Y, REBETS Y, ESTÉVEZ M R, et al. Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters[J]. Microbial Cell Factories, 2020, 19(1): 5. |
| 61 | BALTZ R H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes[J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(2-3): 343-370. |
| 62 | MYRONOVSKYI M, ROSENKRÄNZER B, NADMID S, et al. Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters[J]. Metabolic Engineering, 2018, 49: 316-324. |
| 63 | MUKHERJEE S, STAMATIS D, LI C T, et al. Twenty-five years of Genomes OnLine Database (GOLD): data updates and new features in v.9[J]. Nucleic Acids Research, 2023, 51(D1): D957-D963. |
| 64 | YANG Y L, ZHANG S S, MA K, et al. Discovery and characterization of a new family of diterpene cyclases in bacteria and fungi[J]. Angewandte Chemie International Edition, 2017, 56(17): 4749-4752. |
| 65 | SUN X, CAI Y S, YUAN Y J, et al. Genome mining in Trichoderma viride J1-030: discovery and identification of novel sesquiterpene synthase and its products[J]. Beilstein Journal of Organic Chemistry, 2019, 15: 2052-2058. |
| 66 | CHEN R, FENG T, LI M, et al. Characterization of tremulane sesquiterpene synthase from the basidiomycete Irpex lacteus [J]. Organic Letters, 2022, 24(31): 5669-5673. |
| 67 | LI Z N, XU B F, KOJASOY V, et al. First trans-eunicellane terpene synthase in bacteria[J]. Chem, 2023, 9(3): 698-708. |
| 68 | ZHU C X, XU B F, ADPRESSA D A, et al. Discovery and biosynthesis of a structurally dynamic antibacterial diterpenoid[J]. Angewandte Chemie International Edition, 2021, 60(25): 14163-14170. |
| 69 | WANG Z Y, YANG Q, HE J Y, et al. Cytochrome P450 mediated cyclization in eunicellane derived diterpenoid biosynthesis[J]. Angewandte Chemie International Edition, 2023, 62(45): e202312490. |
| 70 | LI Z, JIANG Y Y, ZHANG X W, et al. Fragrant venezuelaenes A and B with A 5-5-6-7 tetracyclic skeleton: discovery, biosynthesis, and mechanisms of central catalysts[J]. ACS Catalysis, 2020, 10(10): 5846-5851. |
| [1] | WEN Yanhua, LIU Hedong, CAO Chunlai, WU Ruibo. Applications of protein engineering in pharmaceutical industry [J]. Synthetic Biology Journal, 2025, 6(1): 65-86. |
| [2] | SONG Yongxiang, ZHANG Xiufeng, LI Yanqin, XIAO Hua, YAN Yan. Resistance-gene directed discovery of bioactive natural products [J]. Synthetic Biology Journal, 2024, 5(3): 474-491. |
| [3] | XI Mengyu, HU Yiling, GU Yucheng, GE Huiming. Genome mining-directed discovery for natural medicinal products [J]. Synthetic Biology Journal, 2024, 5(3): 447-473. |
| [4] | Jingyong ZHU, Junxiang LI, Xuhui LI, Jin ZHANG, Wenjing WU. Advances in applications of deep learning for predicting sequence-based protein interactions [J]. Synthetic Biology Journal, 2024, 5(1): 88-106. |
| [5] | Fanzhong ZHANG, Changjun XIANG, Lihan ZHANG. Advances and applications of evolutionary analysis and big-data guided bioinformatics in natural product research [J]. Synthetic Biology Journal, 2023, 4(4): 629-650. |
| [6] | Sheng WANG, Zechen WANG, Weihua CHEN, Ke CHEN, Xiangda PENG, Fafen OU, Liangzhen ZHENG, Jinyuan SUN, Tao SHEN, Guoping ZHAO. Design of synthetic biology components based on artificial intelligence and computational biology [J]. Synthetic Biology Journal, 2023, 4(3): 422-443. |
| [7] | Liqi KANG, Pan TAN, Liang HONG. Enzyme engineering in the age of artificial intelligence [J]. Synthetic Biology Journal, 2023, 4(3): 524-534. |
| [8] | Qiaozhen MENG, Fei GUO. Applications of foldability in intelligent enzyme engineering and design: take AlphaFold2 for example [J]. Synthetic Biology Journal, 2023, 4(3): 571-589. |
| [9] | Zhihang CHEN, Menglin JI, Yifei QI. Research progress of artificial intelligence in desiging protein structures [J]. Synthetic Biology Journal, 2023, 4(3): 464-487. |
| [10] | Qilong LAI, Shuai YAO, Yuguo ZHA, Hong BAI, Kang NING. Microbiome-based biosynthetic gene cluster data mining techniques and application potentials [J]. Synthetic Biology Journal, 2023, 4(3): 611-627. |
| [11] | Jiahao BIAN, Guangyu YANG. Artificial intelligence-assisted protein engineering [J]. Synthetic Biology Journal, 2022, 3(3): 429-444. |
| [12] | Qian YANG, Botao CHENG, Zhijun TANG, Wen LIU. Applications and prospects of genome mining in the discovery of natural products [J]. Synthetic Biology Journal, 2021, 2(5): 697-715. |
| [13] | Haibo ZHOU, Qiyao SHEN, Hanna CHEN, Zongjie WANG, Yuezhong LI, Youming ZHANG, Xiaoying BIAN. Genome mining for novel natural products in Sorangium cellulosum So0157-2 by heterologous expression [J]. Synthetic Biology Journal, 2021, 2(5): 837-849. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||