|
||
|
Applications of the CRISPR/Cas9 editing system in the study of microbial natural products
Synthetic Biology Journal
2024, 5 (3):
658-671.
DOI: 10.12211/2096-8280.2023-110
Microorganisms have consistently been a crucial source for researchers to explore and develop new natural products. Currently, research methods involving gene editing tools for the discovery, biosynthesis, and metabolic engineering of natural products have garnered broad attention in this field. However, traditional methods for gene editing usually rely on the recombination ability of the host or introduced proteins. It’s difficult to establish a general platform for all bacteria mainly because of their complicated genetic background. This genetic diversity often causes laborious experimental operations with low efficiency. The CRISPR/Cas9 gene editing system, with its unique and flexible targeting advantages, overcomes common limitations such as sequence homology or site constraint in other gene editing methods and thus is more likely to function in diverse bacteria species. This simplifies experimental procedures, enhances work efficiency, and promotes the development of natural product research. This article introduces the applications of the CRISPR/Cas9 system for the discovery, biosynthesis, and metabolic engineering of natural products in microorganisms. It covers the development of the CRISPR/Cas9 system, cloning and genetic editing of natural product biosynthetic gene clusters, structural derivatization and metabolic engineering of natural products, and the activation of silenced natural product biosynthetic gene clusters. These aspects highlight the advantages of the CRISPR/Cas9 system in the research of natural products with microorganisms. Finally, solutions are proposed for addressing challenges that the CRISPR/Cas9 system currently faces in overcoming low recombination efficiency and host adaptability issues. Especially the CRISPR/Cas12a system which has broadened applications of the CRISRP/Cas9 system by preferring different PAM sites. In addition to functions that CRISPR/Cas9 system has realized, its potent multiple targeting ability further enhances the efficiency of target editing. It is believed that with the development of synthetic biology and information technology, an increasing number of genetic manipulation tools and methods related to the CRISPR/Cas9 system will be developed, continually driving progress in the research of natural products.
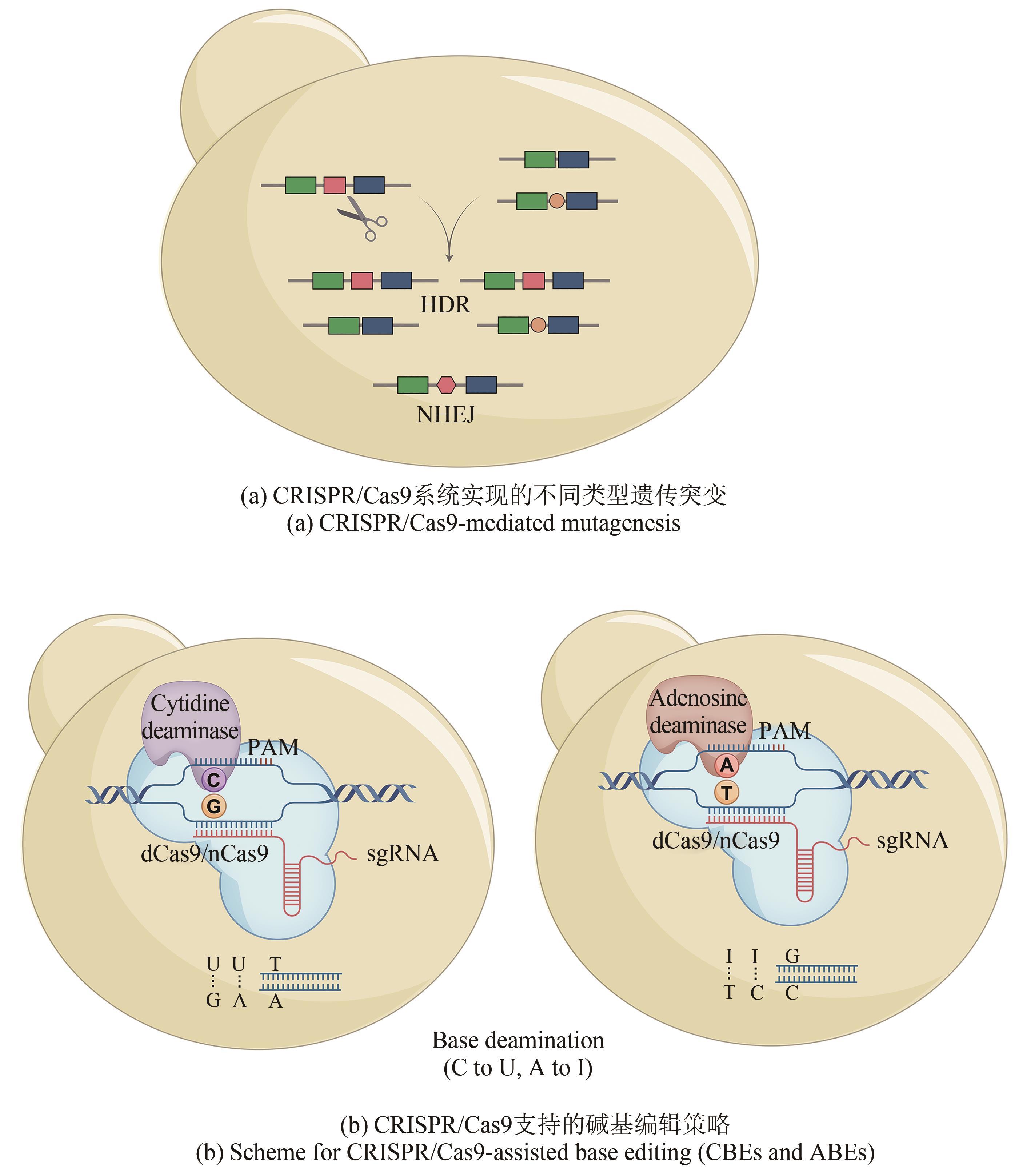
Fig. 2
CRISPR/Cas9-mediated genetic editing of biosynthetic gene clusters
Extracts from the Article
与上面这些较为复杂的基因组编辑手段相比,现在广泛使用的CRISPR/Cas9系统可通过靶向编辑技术直接完成上述复杂重组过程。此外,该系统可实现多位点同时编辑,Cas9蛋白的靶向剪切又能帮助阳性克隆的筛选(表1)。武汉大学孙宇辉团队[34]开发了高效体外ICE(In vitro CRISPR/Cas9-mediated editing)编辑系统,通过额外的T4 DNA连接酶修复体外Cas9/sgRNA反应带来的黏性末端,进而实现了精准无缝的靶向编辑。随后,该团队借助ICE系统分别成功地敲除了来自tetronate RK-682和 dithiolopyrrolone holomycin生物合成基因簇的rkD和homE基因。美国伊利诺伊大学香槟分校(UIUC)赵惠民团队[35]使用温敏型质粒表达来自酿脓链球菌的CRISPR/Cas9系统,并在链霉菌属中实现了多个基因组的靶向编辑。该团队构建的pCRISPomyces质粒可通过Golden Gate组装和等温组装(或传统的消化连接)插入spacer和修复模板,满足快速灵活的基因组编辑。而后通过多重靶向能力成功实现了Streptomyces lividans内31 kb red基因簇的删除。此外,他们还证实该系统适用于多种链霉菌菌株的基因编辑。类似的CRISPR/Cas9介导的基因编辑也被多个团队报道,其中丹麦技术大学Sang Yup Lee团队还证实了在没有引入修复模板时,NHEJ修复通路会在靶向位点周围引入不同大小的缺失突变[36-37,61][图2(a)]。
为实现更快捷的双交联同源重组,在获得含有目的突变克隆的同时去除载体质粒,武汉大学孙宇辉团队[38]通过引入额外的5FC(5-fluorocytosine)反向筛选基因codA(sm),构建了CRISPR/Cas9-CodA(sm)系统。该突变的胞嘧啶脱氨基酶(CodA)可将5FC转换为致死性的毒素5-FU(5-fluorouracil)。通过对来自链霉菌基因组的actI-ORF2的敲除证实了codA(sm)基因带来的反向筛选极大地增加了质粒的清除率和基因敲除率。随后,加州大学圣地亚哥分校的Bradley S. Moore团队[39]开发并借助violacein生物合成基因簇vio验证了CRISPR/Cas9介导的快速点突变能力,他们发现当选择后随链(lagging-strand)和更长的单链DNA为修复模板时同源重组率更高。为了在实现更高突变率的同时省去修复模板,武汉大学孙宇辉团队[40]将CBE和ABE分别与CRISPR/nCas9(high fidelity nickCas9)系统相连,前者完成了对来自S. coelicolor和S. avermitilis的生物合成基因簇的多重位点同时突变,后者通过突变actVB的起始密码子证实了该系统在突变腺嘌呤到鸟嘌呤方面的应用[图2(b)]。此外,中国科学院天津工业生物技术研究所王猛团队[62]借助反义RNA干扰技术同时抑制Streptomyces lividans 66的尿嘧啶DNA糖苷酶的表达,进一步提高了CBE效率。
Other Images/Table from this Article
|