合成生物学 ›› 2023, Vol. 4 ›› Issue (6): 1281-1299.DOI: 10.12211/2096-8280.2023-056
电活性微生物基因编辑与转录调控技术进展与应用
陈雅如1,2, 曹英秀1,2, 宋浩1,2
- 1.天津大学,化工学院,天津 300072
2.天津大学,合成生物学前沿科学中心,系统生物工程教育部重点实验室,天津 300072
-
收稿日期:2023-08-19修回日期:2023-09-18出版日期:2023-12-31发布日期:2024-01-19 -
通讯作者:曹英秀,宋浩 -
作者简介:陈雅如 (1995—),女,博士研究生。研究方向为电活性微生物,基因编辑与调控。E-mail:yaruchen2018207303@tju.edu.cn曹英秀 (1986—),女,副教授,博士生导师。研究方向为高性能生物燃料细胞工厂设计与重构。E-mail:caoyingxiu@tju.edu.cn宋浩 (1973—),男,教授,博士生导师。研究方向为电能细胞合成生物学,微生物光/电合成。E-mail:hsong@tju.edu.cn -
基金资助:国家重点研发计划(2018YFA0901300);国家自然科学基金(32071411)
Advances and applications of gene editing and transcriptional regulation in electroactive microorganisms
CHEN Yaru1,2, CAO Yingxiu1,2, SONG Hao1,2
- 1.School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
2.Key Laboratory of Systems Bioengineering (Ministry of Education),Frontier Science Center for Synthetic Biology,Tianjin University,Tianjin 300072,China
-
Received:2023-08-19Revised:2023-09-18Online:2023-12-31Published:2024-01-19 -
Contact:CAO Yingxiu, SONG Hao
摘要:
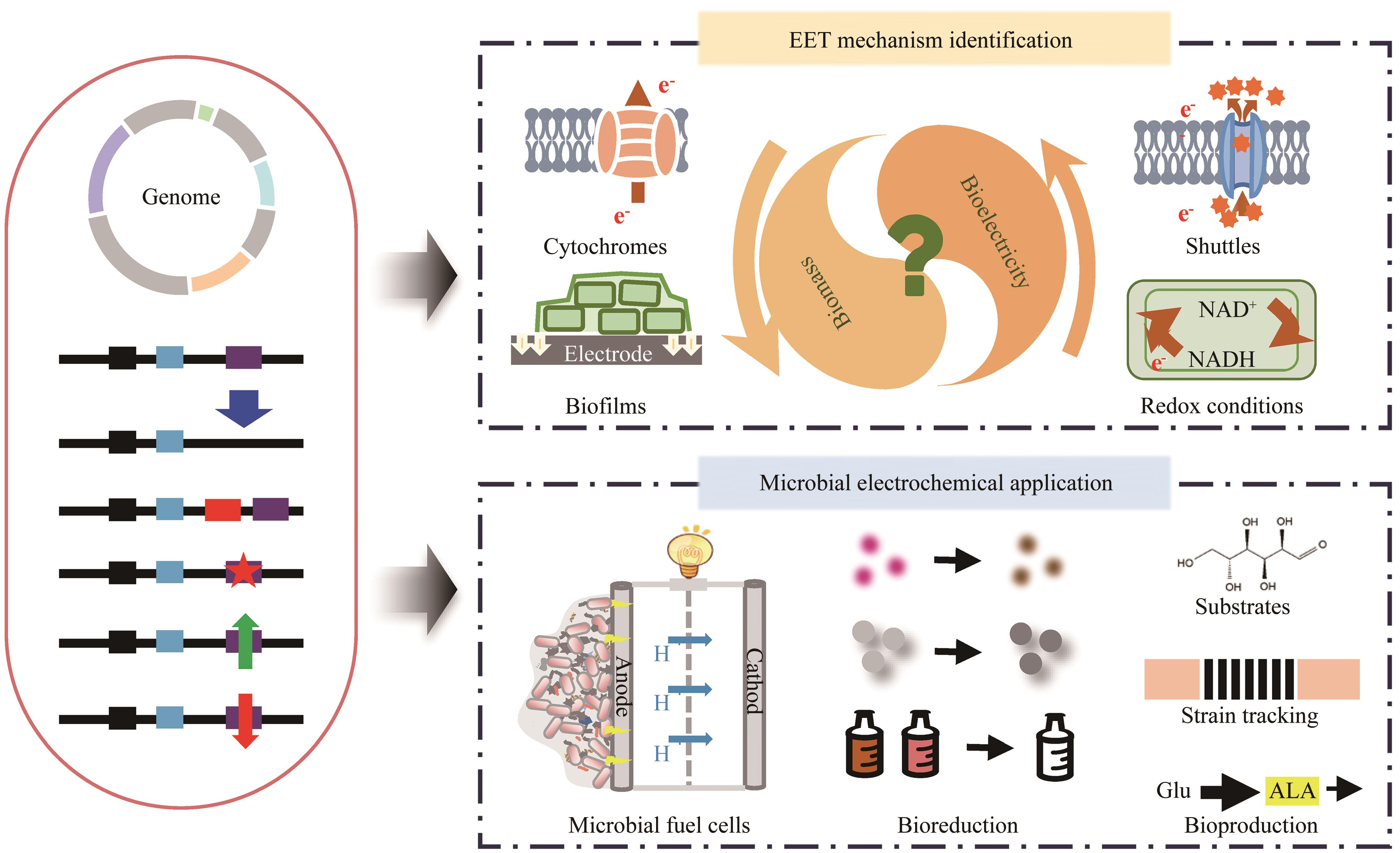
电活性微生物通过胞外电子传递通路与胞外电子受体/供体进行双向电子交换,产生或吞噬电流。电活性微生物已广泛应用于微生物电化学技术领域,涵盖了元素的生物地球化学循环、环境污染的生物处理与电能生产、生物传感、微生物冶金以及化学品的微生物电合成等多个领域,成为全球环境保护和低碳经济的研究热点。然而,这些微生物在实际应用中仍面临较大局限,如微生物燃料电池的输出功率密度存在一定的上限、微生物电合成技术中的CO2还原速率尚未达到理想水平等。为了克服这些限制性因素,需要通过高效的基因编辑和转录调控策略来改变电活性微生物的遗传特性,提高其双向电子传递效率。本文首先总结了模式电活性微生物(希瓦氏菌和地杆菌)和其他代表性电活性微生物的基因编辑方法和利用CRISPR(clustered regularly interspaced short palindromic repeat)技术实现转录调控的策略。在基因编辑方面,涵盖了(CRISPR辅助的)同源重组、碱基编辑等方法;而在转录调控方面,包括了CRISPR介导的抑制和激活。此外,对于多基因编辑和调控的策略也进行了深入探讨。其次,综述了这些技术在环境、能源领域中的应用,包括微生物燃料电池、污染物生物处理和修复等。最后,讨论了目前电活性微生物工程改造所面临的挑战和未来的发展方向。
中图分类号:
引用本文
陈雅如, 曹英秀, 宋浩. 电活性微生物基因编辑与转录调控技术进展与应用[J]. 合成生物学, 2023, 4(6): 1281-1299.
CHEN Yaru, CAO Yingxiu, SONG Hao. Advances and applications of gene editing and transcriptional regulation in electroactive microorganisms[J]. Synthetic Biology Journal, 2023, 4(6): 1281-1299.
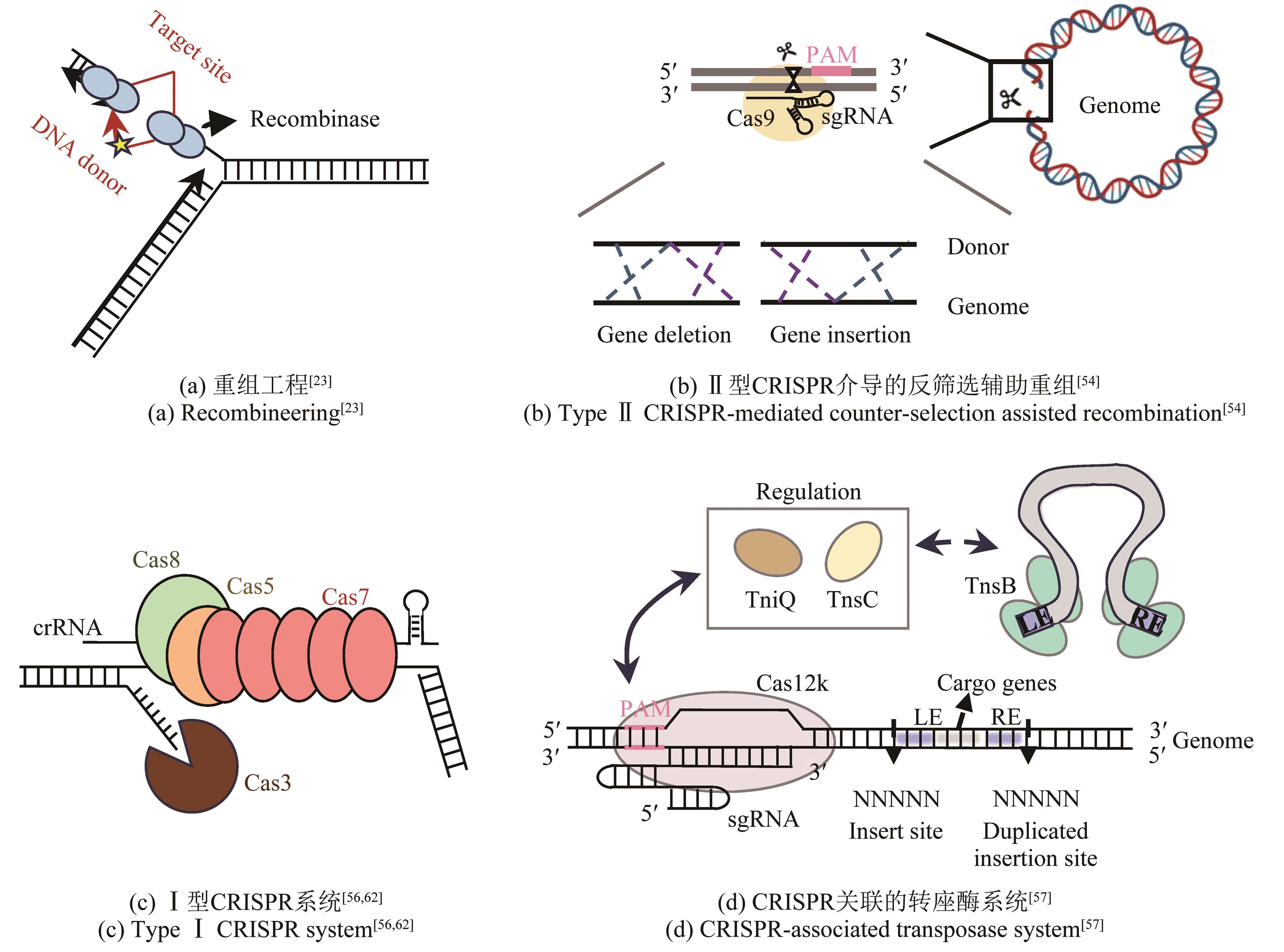
| 1 | LOVLEY D R, HOLMES D E. Electromicrobiology: the ecophysiology of phylogenetically diverse electroactive microorganisms[J]. Nature Reviews Microbiology, 2022, 20(1): 5-19. |
| 2 | JIANG Y, ZENG R J. Bidirectional extracellular electron transfers of electrode-biofilm: mechanism and application[J]. Bioresource Technology, 2019, 271: 439-448. |
| 3 | LOGAN B E, ROSSI R, RAGAB A, et al. Electroactive microorganisms in bioelectrochemical systems[J]. Nature Reviews Microbiology, 2019, 17(5): 307-319. |
| 4 | CHU N, LIANG Q J, HAO W, et al. Microbial electrochemical sensor for water biotoxicity monitoring[J]. Chemical Engineering Journal, 2021, 404: 127053. |
| 5 | LIU C J, YU H, ZHANG B C, et al. Engineering whole-cell microbial biosensors: design principles and applications in monitoring and treatment of heavy metals and organic pollutants[J]. Biotechnology Advances, 2022, 60: 108019. |
| 6 | LÜ J, REN G P, HU Q C, et al. Microbial biofilm-based hydrovoltaic technology[J]. Trends in Biotechnology, 2023, 41(9): 1155-1167. |
| 7 | LOVLEY D R, HOLMES D E. Protein nanowires: the electrification of the microbial world and maybe our own[J]. Journal of Bacteriology, 2020, 202(20): e00331-20. |
| 8 | CHEN H, DONG F Y, MINTEER S D. The progress and outlook of bioelectrocatalysis for the production of chemicals, fuels and materials[J]. Nature Catalysis, 2020, 3(3): 225-244. |
| 9 | CESTELLOS-BLANCO S, ZHANG H, KIM J M, et al. Photosynthetic semiconductor biohybrids for solar-driven biocatalysis[J]. Nature Catalysis, 2020, 3(3): 245-255. |
| 10 | CAO B C, ZHAO Z P, PENG L L, et al. Silver nanoparticles boost charge-extraction efficiency in Shewanella microbial fuel cells[J]. Science, 2021, 373(6561): 1336-1340. |
| 11 | YU Y Y, WANG Y Z, FANG Z, et al. Single cell electron collectors for highly efficient wiring-up electronic abiotic/biotic interfaces[J]. Nature Communications, 2020, 11: 4087. |
| 12 | TABARES M, DULAY H, REGUERA G. Geobacter sulfurreducens[J]. Trends in Microbiology, 2020, 28(4): 327-328. |
| 13 | MENG F K, ELLIS T. The second decade of synthetic biology: 2010-2020[J]. Nature Communications, 2020, 11: 5174. |
| 14 | SHARAN S K, THOMASON L C, KUZNETSOV S G, et al. Recombineering: a homologous recombination-based method of genetic engineering[J]. Nature Protocols, 2009, 4(2): 206-223. |
| 15 | FAN Y Y, TANG Q A, LI Y, et al. Rapid and highly efficient genomic engineering with a novel iEditing device for programming versatile extracellular electron transfer of electroactive bacteria[J]. Environmental Microbiology, 2021, 23(2): 1238-1255. |
| 16 | COPPI M V, LEANG C, SANDLER S J, et al. Development of a genetic system for Geobacter sulfurreducens [J]. Applied and Environmental Microbiology, 2001, 67(7): 3180-3187. |
| 17 | CHEN W Z, ZHANG Y, ZHANG Y F, et al. CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species[J]. iScience, 2018, 6: 222-231. |
| 18 | LI Y, LI Y Y, CHEN Y R, et al. Coupling riboflavin de novo biosynthesis and cytochrome expression for improving extracellular electron transfer efficiency in Shewanella oneidensis [J]. Biotechnology and Bioengineering, 2022, 119(10): 2806-2818. |
| 19 | WU Z Y, HUANG Y T, CHAO W C, et al. Reversal of carbapenem-resistance in Shewanella algae by CRISPR/Cas9 genome editing[J]. Journal of Advanced Research, 2019, 18: 61-69. |
| 20 | LIU T, YU Y Y, DENG X P, et al. Enhanced Shewanella biofilm promotes bioelectricity generation[J]. Biotechnology and Bioengineering, 2015, 112(10): 2051-2059. |
| 21 | LI F, LI Y X, CAO Y X, et al. Modular engineering to increase intracellular NAD(H/+) promotes rate of extracellular electron transfer of Shewanella oneidensis [J]. Nature Communications, 2018, 9: 3637. |
| 22 | ZHENG W T, XIA Y D, WANG X E, et al. Precise genome engineering in Pseudomonas using phage-encoded homologous recombination and the Cascade-Cas3 system[J]. Nature Protocols, 2023, 18(9): 2642-2670. |
| 23 | CORTS A D, THOMASON L C, GILL R T, et al. A new recombineering system for precise genome-editing in Shewanella oneidensis strain MR-1 using single-stranded oligonucleotides[J]. Scientific Reports, 2019, 9: 39. |
| 24 | HMELO L R, BORLEE B R, ALMBLAD H, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange[J]. Nature Protocols, 2015, 10(11): 1820-1841. |
| 25 | THORMANN K M, SAVILLE R M, SHUKLA S, et al. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms[J]. Journal of Bacteriology, 2005, 187(3): 1014-1021. |
| 26 | CHAN C H, LEVAR C E, ZACHAROFF L, et al. Scarless genome editing and stable inducible expression vectors for Geobacter sulfurreducens [J]. Applied and Environmental Microbiology, 2015, 81(20): 7178-7186. |
| 27 | GRAF N, ALTENBUCHNER J. Development of a method for markerless gene deletion in Pseudomonas putida [J]. Applied and Environmental Microbiology, 2011, 77(15): 5549-5552. |
| 28 | PÓSFAI G, KOLISNYCHENKO V, BERECZKI Z, et al. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome[J]. Nucleic Acids Research, 1999, 27(22): 4409-4415. |
| 29 | MARTÍNEZ-GARCÍA E, DE LORENZO V. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440[J]. Environmental Microbiology, 2011, 13(10): 2702-2716. |
| 30 | LUO X, YANG Y W, LING W, et al. Pseudomonas putida KT2440 markerless gene deletion using a combination of λ Red recombineering and Cre/loxP site-specific recombination[J]. FEMS Microbiology Letters, 2016, 363(4): fnw014. |
| 31 | VELÁZQUEZ E, AL-RAMAHI Y, TELLECHEA-LUZARDO J, et al. Targetron-assisted delivery of exogenous DNA sequences into Pseudomonas putida through CRISPR-aided counterselection[J]. ACS Synthetic Biology, 2021, 10(10): 2552-2565. |
| 32 | BIRD L J, KUNDU B B, TSCHIRHART T, et al. Engineering wired life: synthetic biology for electroactive bacteria[J]. ACS Synthetic Biology, 2021, 10(11): 2808-2823. |
| 33 | MURPHY K C. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli [J]. Journal of Bacteriology, 1998, 180(8): 2063-2071. |
| 34 | SAWITZKE J A, THOMASON L C, COSTANTINO N, et al. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond[J]. Methods in Enzymology, 2007, 421: 171-199. |
| 35 | PINES G, FREED E F, WINKLER J D, et al. Bacterial recombineering: genome engineering via phage-based homologous recombination[J]. ACS Synthetic Biology, 2015, 4(11): 1176-1185. |
| 36 | SWINGLE B, BAO Z M, MARKEL E, et al. Recombineering using RecTE from Pseudomonas syringae [J]. Applied and Environmental Microbiology, 2010, 76(15): 4960-4968. |
| 37 | BAO Z M, CARTINHOUR S, SWINGLE B. Substrate and target sequence length influence RecTEPsy recombineering efficiency in Pseudomonas syringae [J]. PLoS One, 2012, 7(11): e50617. |
| 38 | APARICIO T, JENSEN S I, NIELSEN A T, et al. The Ssr protein (T1E_1405) from Pseudomonas putida DOT-T1E enables oligonucleotide-based recombineering in platform strain P. putida EM42[J]. Biotechnology Journal, 2016, 11(10): 1309-1319. |
| 39 | WANG H H, ISAACS F J, CARR P A, et al. Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894-898. |
| 40 | CARROLL D. Genome engineering with targetable nucleases[J]. Annual Review of Biochemistry, 2014, 83: 409-439. |
| 41 | RAN F A, HSU P D, WRIGHT J, et al. Genome engineering using the CRISPR-Cas9 system[J]. Nature Protocols, 2013, 8(11): 2281-2308. |
| 42 | STERNBERG S H, REDDING S, JINEK M, et al. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9[J]. Nature, 2014, 507(7490): 62-67. |
| 43 | PETERS J M, COLAVIN A, SHI H D, et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria[J]. Cell, 2016, 165(6): 1493-1506. |
| 44 | FAN Y Y, TANG Q A, LI F H, et al. Enhanced bioreduction of radionuclides by driving microbial extracellular electron pumping with an engineered CRISPR platform[J]. Environmental Science & Technology, 2021, 55(17): 11997-12008. |
| 45 | WIRTH N T, KOZAEVA E, NIKEL P I. Accelerated genome engineering of Pseudomonas putida by Ⅰ-SceⅠ-mediated recombination and CRISPR-Cas9 counterselection[J]. Microbial Biotechnology, 2020, 13(1): 233-249. |
| 46 | CORTS A D, THOMASON L C, GILL R T, et al. Efficient and precise genome editing in Shewanella with recombineering and CRISPR/Cas9-mediated counter-selection[J]. ACS Synthetic Biology, 2019, 8(8): 1877-1889. |
| 47 | COOK T B, RAND J M, NURANI W, et al. Genetic tools for reliable gene expression and recombineering in Pseudomonas putida [J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(7): 517-527. |
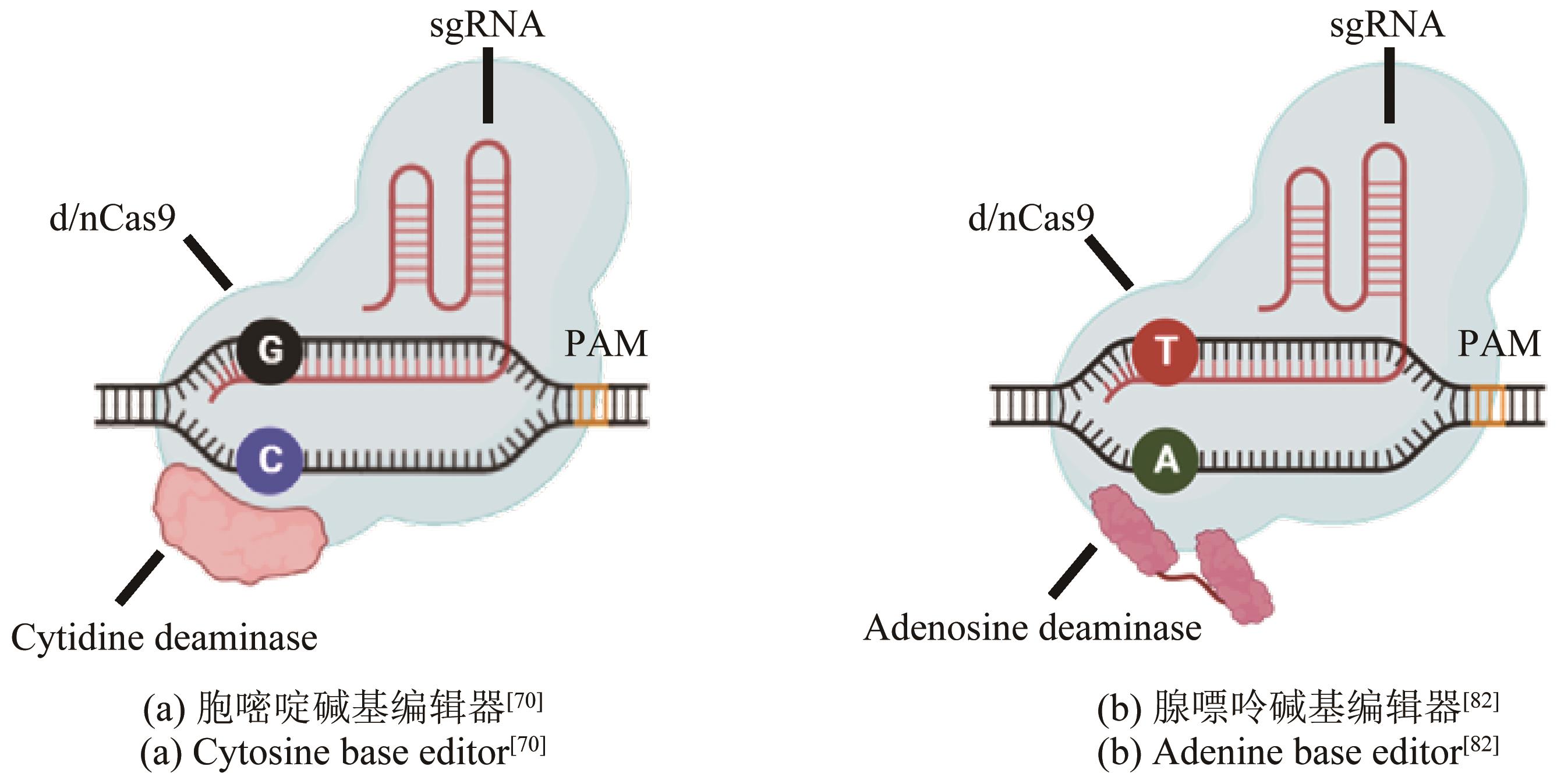
| 48 | WU Z X, CHEN Z Q, GAO X Y, et al. Combination of ssDNA recombineering and CRISPR-Cas9 for Pseudomonas putida KT2440 genome editing[J]. Applied Microbiology and Biotechnology, 2019, 103(6): 2783-2795. |
| 49 | ZHOU Y Y, LIN L, WANG H, et al. Development of a CRISPR/Cas9n-based tool for metabolic engineering of Pseudomonas putida for ferulic acid-to-polyhydroxyalkanoate bioconversion[J]. Communications Biology, 2020, 3: 98. |
| 50 | SUN J, WANG Q Z, JIANG Y, et al. Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type Ⅱ CRISPR system[J]. Microbial Cell Factories, 2018, 17(1): 41. |
| 51 | GARST A D, BASSALO M C, PINES G, et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering[J]. Nature Biotechnology, 2017, 35(1): 48-55. |
| 52 | PETERS J E, MAKAROVA K S, SHMAKOV S, et al. Recruitment of CRISPR-Cas systems by Tn7-like transposons[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(35): E7358-E7366. |
| 53 | ASIN-GARCIA E, MARTIN-PASCUAL M, GARCIA-MORALES L, et al. ReScribe: an unrestrained tool combining multiplex recombineering and minimal-PAM ScCas9 for genome recoding Pseudomonas putida [J]. ACS Synthetic Biology, 2021, 10(10): 2672-2688. |
| 54 | CHEN Y R, CHENG M J, FENG X R, et al. Genome editing by CRISPR/Cas12 recognizing AT-rich PAMs in Shewanella oneidensis MR-1[J]. ACS Synthetic Biology, 2022, 11(9): 2947-2955. |
| 55 | LIN Z L, LI H H, HE L, et al. Efficient genome editing for Pseudomonas aeruginosa using CRISPR-Cas12a[J]. Gene, 2021, 790: 145693. |
| 56 | CSÖRGŐ B, LEÓN L M, CHAU-LY I J, et al. A compact Cascade-Cas3 system for targeted genome engineering[J]. Nature Methods, 2020, 17(12): 1183-1190. |
| 57 | CHENG Z H, WU J, LIU J Q, et al. Repurposing CRISPR RNA-guided integrases system for one-step, efficient genomic integration of ultra-long DNA sequences[J]. Nucleic Acids Research, 2022, 50(13): 7739-7750. |
| 58 | MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants[J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. |
| 59 | XU Z L, LI M, LI Y R, et al. Native CRISPR-cas-mediated genome editing enables dissecting and sensitizing clinical multidrug-resistant P. aeruginosa [J]. Cell Reports, 2019, 29(6): 1707-1717.e3. |
| 60 | DWARAKANATH S, BRENZINGER S, GLEDITZSCH D, et al. Interference activity of a minimal TypeⅠCRISPR-Cas system from Shewanella putrefaciens [J]. Nucleic Acids Research, 2015, 43(18): 8913-8923. |
| 61 | CHEN Y R, CHENG M J, SONG H, et al. Type Ⅰ-F CRISPR-PAIR platform for multi-mode regulation to boost extracellular electron transfer in Shewanella oneidensis [J]. iScience, 2022, 25(6): 104491. |
| 62 | CHEN Y X, LIU J Q, ZHI S Y, et al. Repurposing type Ⅰ-F CRISPR-Cas system as a transcriptional activation tool in human cells[J]. Nature Communications, 2020, 11: 3136. |
| 63 | XU Z L, LI Y R, CAO H L, et al. A transferrable and integrative type Ⅰ-F Cascade for heterologous genome editing and transcription modulation[J]. Nucleic Acids Research, 2021, 49(16): e94. |
| 64 | KLOMPE S E, VO P L H, HALPIN-HEALY T S, et al. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration[J]. Nature, 2019, 571(7764): 219-225. |
| 65 | STRECKER J, LADHA A, GARDNER Z, et al. RNA-guided DNA insertion with CRISPR-associated transposases[J]. Science, 2019, 365(6448): 48-53. |
| 66 | WANG Y, LIU Y, ZHENG P, et al. Microbial base editing: a powerful emerging technology for microbial genome engineering[J]. Trends in Biotechnology, 2021, 39(2): 165-180. |
| 67 | CHENG L, MIN D, HE R L, et al. Developing a base-editing system to expand the carbon source utilization spectra of Shewanella oneidensis MR-1 for enhanced pollutant degradation[J]. Biotechnology and Bioengineering, 2020, 117(8): 2389-2400. |
| 68 | ABDULLAH, WANG P J, HAN T R, et al. Adenine base editing system for Pseudomonas and prediction workflow for protein dysfunction via ABE[J]. ACS Synthetic Biology, 2022, 11(4): 1650-1657. |
| 69 | MOLLA K A, YANG Y N. CRISPR/cas-mediated base editing: technical considerations and practical applications[J]. Trends in Biotechnology, 2019, 37(10): 1121-1142. |
| 70 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
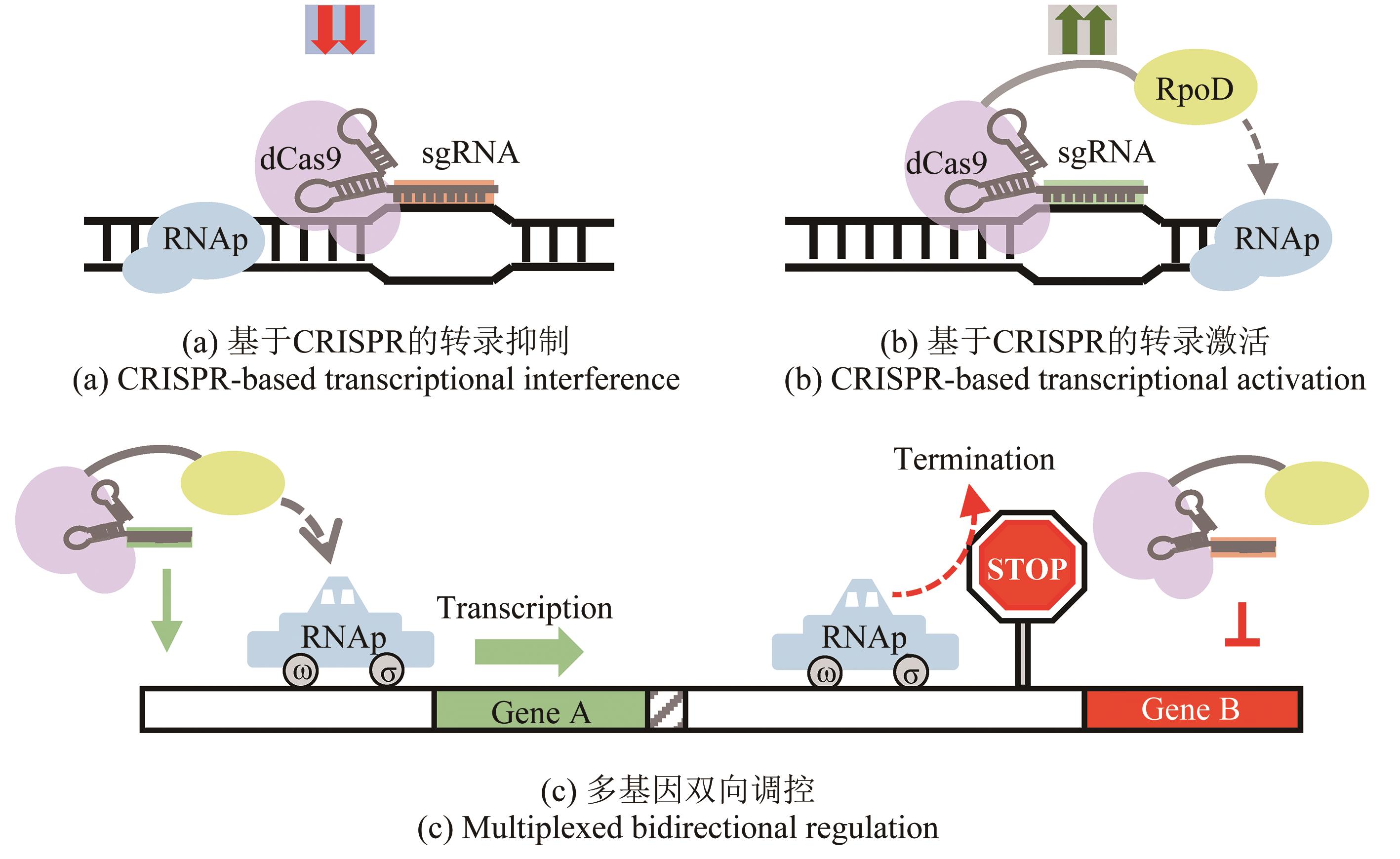
| 71 | NISHIDA K, ARAZOE T, YACHIE N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems[J]. Science, 2016, 353(6305): aaf8729. |
| 72 | KUSCU C, PARLAK M, TUFAN T R, et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations[J]. Nature Methods, 2017, 14(7): 710-712. |
| 73 | WANG Y, LIU Y, LIU J, et al. MACBETH: multiplex automated Corynebacterium glutamicum base editing method[J]. Metabolic Engineering, 2018, 47: 200-210. |
| 74 | CHEN Y R, FANG L X, YING X A, et al. Development of whole genome-scale base editing toolbox to promote efficiency of extracellular electron transfer in Shewanella oneidensis MR-1[J]. Advanced Biology, 2022, 6(3): 2101296. |
| 75 | HE R L, WU J E, CHENG Z H, et al. Biomolecular insights into extracellular pollutant reduction pathways of Geobacter sulfurreducens using a base editor system[J]. Environmental Science & Technology, 2022, 56(17): 12247-12256. |
| 76 | YUE S J, HUANG P, LI S, et al. Developing a CRISPR-assisted base-editing system for genome engineering of Pseudomonas chlororaphis [J]. Microbial Biotechnology, 2022, 15(9): 2324-2336. |
| 77 | SUN J, LU L B, LIANG T X, et al. CRISPR-assisted multiplex base editing system in Pseudomonas putida KT2440[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 905. |
| 78 | WEGRZYN R D, LEE A H, JENKINS A L, et al. Genome editing: insights from chemical biology to support safe and transformative therapeutic applications[J]. ACS Chemical Biology, 2018, 13(2): 333-342. |
| 79 | JAIN S, XUN G H, ABESTEH S, et al. Precise regulation of Cas9-mediated genome engineering by anti-CRISPR-based inducible CRISPR controllers[J]. ACS Synthetic Biology, 2021, 10(6): 1320-1327. |
| 80 | SAITO M, XU P Y, FAURE G, et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease[J]. Nature, 2023, 620(7974): 660-668. |
| 81 | MATSOUKAS I G. Commentary: programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Frontiers in Genetics, 2018, 9: 21. |
| 82 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| 83 | WANG X J, LIU Z W, LI G L, et al. Efficient gene silencing by adenine base editor-mediated start codon mutation[J]. Molecular Therapy, 2020, 28(2): 431-440. |
| 84 | ZHANG Y, ZHANG H Y, WANG Z P, et al. Programmable adenine deamination in bacteria using a Cas9-adenine-deaminase fusion[J]. Chemical Science, 2020, 11(6): 1657-1664. |
| 85 | WANG T L, ZHANG J W, WEI L, et al. Developing a PAM-flexible CRISPR-mediated dual-deaminase base editor to regulate extracellular electron transport in Shewanella oneidensis [J]. ACS Synthetic Biology, 2023, 12(6): 1727-1738. |
| 86 | LI C, ZHANG R, MENG X B, et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors[J]. Nature Biotechnology, 2020, 38(7): 875-882. |
| 87 | APARICIO T, DE LORENZO V, MARTÍNEZ-GARCÍA E. CRISPR/Cas9-enhanced ssDNA recombineering for Pseudomonas putida [J]. Microbial Biotechnology, 2019, 12(5): 1076-1089. |
| 88 | MCCARTY N S, GRAHAM A E, STUDENÁ L, et al. Multiplexed CRISPR technologies for gene editing and transcriptional regulation[J]. Nature Communications, 2020, 11: 1281. |
| 89 | CHEN Y R, CHENG M J, LI Y, et al. Highly efficient multiplex base editing: one-shot deactivation of eight genes in Shewanella oneidensis MR-1[J]. Synthetic and Systems Biotechnology, 2023, 8(1): 1-10. |
| 90 | VOLKE D C, MARTINO R A, KOZAEVA E, et al. Modular (de)construction of complex bacterial phenotypes by CRISPR/nCas9-assisted, multiplex cytidine base-editing[J]. Nature Communications, 2022, 13: 3026. |
| 91 | WU J, CHENG Z H, MIN D, et al. CRISPRi system as an efficient, simple platform for rapid identification of genes involved in pollutant transformation by Aeromonas hydrophila [J]. Environmental Science & Technology, 2020, 54(6): 3306-3315. |
| 92 | CAO Y X, LI X F, LI F, et al. CRISPRi-sRNA: transcriptional-translational regulation of extracellular electron transfer in Shewanella oneidensis [J]. ACS Synthetic Biology, 2017, 6(9): 1679-1690. |
| 93 | CHEN Y R, NIU X L, CHENG M J, et al. CRISPR/dCas9-RpoD-mediated simultaneous transcriptional activation and repression in Shewanella oneidensis MR-1[J]. ACS Synthetic Biology, 2022, 11(6): 2184-2192. |
| 94 | LARSON M H, GILBERT L A, WANG X W, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression[J]. Nature Protocols, 2013, 8(11): 2180-2196. |
| 95 | LI J E, TANG Q A, LI Y, et al. Rediverting electron flux with an engineered CRISPR-ddAsCpf1 system to enhance the pollutant degradation capacity of Shewanella oneidensis [J]. Environmental Science & Technology, 2020, 54(6): 3599-3608. |
| 96 | QI L S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression[J]. Cell, 2013, 152(5): 1173-1183. |
| 97 | LIU Y, WAN X Y, WANG B J. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria[J]. Nature Communications, 2019, 10: 3693. |
| 98 | BROWNING D F, BUSBY S J W. Local and global regulation of transcription initiation in bacteria[J]. Nature Reviews Microbiology, 2016, 14(10): 638-650. |
| 99 | TANENBAUM M E, GILBERT L A, QI L S, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging[J]. Cell, 2014, 159(3): 635-646. |
| 100 | HO H I, FANG J R, CHEUNG J, et al. Programmable CRISPR-Cas transcriptional activation in bacteria[J]. Molecular Systems Biology, 2020, 16(7): e9427. |
| 101 | CZAJKA J J, BANERJEE D, ENG T, et al. Tuning a high performing multiplexed-CRISPRi Pseudomonas putida strain to further enhance indigoidine production[J]. Metabolic Engineering Communications, 2022, 15: e00206. |
| 102 | LI F, TANG R, ZHANG B C, et al. Systematic full-cycle engineering microbial biofilms to boost electricity production in Shewanella oneidensis [J]. Research, 2023, 6: 81. |
| 103 | BHOKISHAM N, VANARSDALE E, STEPHENS K T, et al. A redox-based electrogenetic CRISPR system to connect with and control biological information networks[J]. Nature Communications, 2020, 11: 2427. |
| 104 | SCHUETZ B, SCHICKLBERGER M, KUERMANN J, et al. Periplasmic electron transfer via the c-type cytochromes MtrA and FccA of Shewanella oneidensis MR-1[J]. Applied and Environmental Microbiology, 2009, 75(24): 7789-7796. |
| 105 | YU H, LU Y J, LAN F, et al. Engineering outer membrane vesicles to increase extracellular electron transfer of Shewanella oneidensis [J]. ACS Synthetic Biology, 2023, 12(6): 1645-1656. |
| 106 | SHI L, DONG H L, REGUERA G, et al. Extracellular electron transfer mechanisms between microorganisms and minerals[J]. Nature Reviews Microbiology, 2016, 14(10): 651-662. |
| 107 | THANASSI D G, BLISKA J B, CHRISTIE P J. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function[J]. FEMS Microbiology Reviews, 2012, 36(6): 1046-1082. |
| 108 | WANG F B, GU Y Q, O'BRIEN J P, et al. Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers[J]. Cell, 2019, 177(2): 361-369.e10. |
| 109 | YALCIN S E, MALVANKAR N S. The blind men and the filament: understanding structures and functions of microbial nanowires[J]. Current Opinion in Chemical Biology, 2020, 59: 193-201. |
| 110 | LOVLEY D R, WALKER D J F. Geobacter protein nanowires[J]. Frontiers in Microbiology, 2019, 10: 2078. |
| 111 | LEANG C, MALVANKAR N S, FRANKS A E, et al. Engineering Geobacter sulfurreducens to produce a highly cohesive conductive matrix with enhanced capacity for current production[J]. Energy & Environmental Science, 2013, 6(6): 1901-1908. |
| 112 | PIRBADIAN S, BARCHINGER S E, LEUNG K M, et al. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(35): 12883-12888. |
| 113 | CATANIA C, KARBELKAR A A, FURST A L. Engineering the interface between electroactive bacteria and electrodes[J]. Joule, 2021, 5(4): 743-747. |
| 114 | HEIJNE A TER, PEREIRA M A, PEREIRA J, et al. Electron storage in electroactive biofilms[J]. Trends in Biotechnology, 2021, 39(1): 34-42. |
| 115 | STURM G, RICHTER K, DOETSCH A, et al. A dynamic periplasmic electron transfer network enables respiratory flexibility beyond a thermodynamic regulatory regime[J]. The ISME Journal, 2015, 9(8): 1802-1811. |
| 116 | FANG L X, LI Y Y, LI Y, et al. Transcriptome analysis to identify crucial genes for reinforcing flavins-mediated extracellular electron transfer in Shewanella oneidensis [J]. Frontiers in Microbiology, 2022, 13: 852527. |
| 117 | MIN D, CHENG L, ZHANG F, et al. Enhancing extracellular electron transfer of Shewanella oneidensis MR-1 through coupling improved flavin synthesis and metal-reducing conduit for pollutant degradation[J]. Environmental Science & Technology, 2017, 51(9): 5082-5089. |
| 118 | YI Y C, NG I S. Redirection of metabolic flux in Shewanella oneidensis MR-1 by CRISPRi and modular design for 5-aminolevulinic acid production[J]. Bioresources and Bioprocessing, 2021, 8: 13. |
| 119 | SCHUSTER A, ERASIMUS H, FRITAH S, et al. RNAi/CRISPR screens: from a pool to a valid hit[J]. Trends in Biotechnology, 2019, 37(1): 38-55. |
| 120 | ALPER H, MOXLEY J, NEVOIGT E, et al. Engineering yeast transcription machinery for improved ethanol tolerance and production[J]. Science, 2006, 314(5805): 1565-1568. |
| 121 | WARNER J R, REEDER P J, KARIMPOUR-FARD A, et al. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides[J]. Nature Biotechnology, 2010, 28(8): 856-862. |
| 122 | BAO Z H, HAMEDIRAD M, XUE P, et al. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision[J]. Nature Biotechnology, 2018, 36(6): 505-508. |
| 123 | YAO L, SHABESTARY K, BJÖRK S M, et al. Pooled CRISPRi screening of the cyanobacterium Synechocystis sp. PCC 6803 for enhanced industrial phenotypes[J]. Nature Communications, 2020, 11: 1666. |
| 124 | WANG T M, GUAN C G, GUO J H, et al. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance[J]. Nature Communications, 2018, 9: 2475. |
| 125 | LIAN J Z, SCHULTZ C, CAO M F, et al. Multi-functional genome-wide CRISPR system for high throughput genotype-phenotype mapping[J]. Nature Communications, 2019, 10: 5794. |
| 126 | XIAO X, LIU Q Y, LI T T, et al. A high-throughput dye-reducing photometric assay for evaluating microbial exoelectrogenic ability[J]. Bioresource Technology, 2017, 241: 743-749. |
| 127 | YANG Z C, CHENG Y Y, ZHANG F, et al. Rapid detection and enumeration of exoelectrogenic bacteria in lake sediments and a wastewater treatment plant using a coupled WO3 nanoclusters and most probable number method[J]. Environmental Science & Technology Letters, 2016, 3(4): 133-137. |
| [1] | 董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117. |
| [2] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [3] | 陈盈盈, 刘扬, 史俊杰, 马俊英, 鞠建华. CRISPR/Cas基因编辑及其新兴技术在丝状真菌研究中的系统应用[J]. 合成生物学, 2024, 5(3): 672-693. |
| [4] | 惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671. |
| [5] | 杜瑶, 高宏丹, 刘家坤, 刘孝荣, 邢志浩, 张涛, 马东礼. CRISPR-Cas系统在病原核酸检测中的研究进展[J]. 合成生物学, 2024, 5(1): 202-216. |
| [6] | 许志锰, 谢震. 引导编辑研究进展及其应用[J]. 合成生物学, 2024, 5(1): 1-15. |
| [7] | 马孟丹, 尚梦宇, 刘宇辰. CRISPR-Cas9系统在肿瘤生物学中的应用及前景[J]. 合成生物学, 2023, 4(4): 703-719. |
| [8] | 王甜甜, 朱虹, 杨琛. 蓝细菌CRISPRa系统的开发及其代谢工程应用[J]. 合成生物学, 2023, 4(4): 824-839. |
| [9] | 林继聪, 邹根, 刘宏民, 魏勇军. CRISPR/Cas基因组编辑技术在丝状真菌次级代谢产物合成中的应用[J]. 合成生物学, 2023, 4(4): 738-755. |
| [10] | 马孟丹, 刘宇辰. 合成生物学在疾病信息记录与实时监测中的应用潜力[J]. 合成生物学, 2023, 4(2): 301-317. |
| [11] | 柳柯, 林桂虹, 刘坤, 周伟, 王风清, 魏东芝. CRISPR/Cas系统的挖掘、改造与功能拓展[J]. 合成生物学, 2023, 4(1): 47-66. |
| [12] | 滕小龙, 史硕博. CRISPR/Cas9系统在基因组编辑中的优化与发展[J]. 合成生物学, 2023, 4(1): 67-85. |
| [13] | 梁丽亚, 刘嵘明. 靶向DNA的Ⅱ类CRISPR/Cas系统的蛋白工程化改造[J]. 合成生物学, 2023, 4(1): 86-101. |
| [14] | 潘颖佳, 夏思杨, 董昌, 蔡谨, 连佳长. 基因增变器驱动的酿酒酵母基因组连续进化[J]. 合成生物学, 2023, 4(1): 225-240. |
| [15] | 刘佳昕, 程驰, 李欣启, 汪超俊, 张颖, 薛闯. 梭菌分子遗传改造工具研究进展[J]. 合成生物学, 2022, 3(6): 1201-1217. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||