合成生物学 ›› 2023, Vol. 4 ›› Issue (3): 524-534.DOI: 10.12211/2096-8280.2023-009
人工智能时代下的酶工程
康里奇1,2, 谈攀3, 洪亮1,2
- 1.上海交通大学物理与天文学院,上海 200240
2.上海交通大学自然科学研究院,上海国家应用数学中心(交大分中心),上海 200240
3.上海人工智能实验室,上海 200240
-
收稿日期:2023-01-16修回日期:2023-03-29出版日期:2023-06-30发布日期:2023-07-05 -
通讯作者:谈攀,洪亮 -
作者简介:康里奇 (1997—),男,博士研究生。研究方向为人工智能功能蛋白质设计。 E-mail:liqikang@sjtu.edu.cn谈攀 (1990—),男,博士,研究员。研究方向为分子生物物理,人工智能功能蛋白质设计以及药物分子设计。 E-mail:tpan1039@sjtu.edu.cn洪亮 (1981—),男,教授,博士生导师。研究方向为分子生物物理,人工智能功能蛋白质设计以及药物分子设计。 E-mail:hongl3liang@sjtu.edu.cn -
基金资助:国家自然科学基金(12104295)
Enzyme engineering in the age of artificial intelligence
KANG Liqi1,2, TAN Pan3, HONG Liang1,2
- 1.School of Physics and Astronomy,Shanghai Jiao Tong University,Shanghai 200240,China
2.Shanghai National Center for Applied Mathematics (SJTU Center),Institute of Natural Sciences,Shanghai Jiao Tong University,Shanghai 200240,China
3.Shanghai Artificial Intelligence Laboratory,Shanghai 200240,China
-
Received:2023-01-16Revised:2023-03-29Online:2023-06-30Published:2023-07-05 -
Contact:TAN Pan, HONG Liang
摘要:
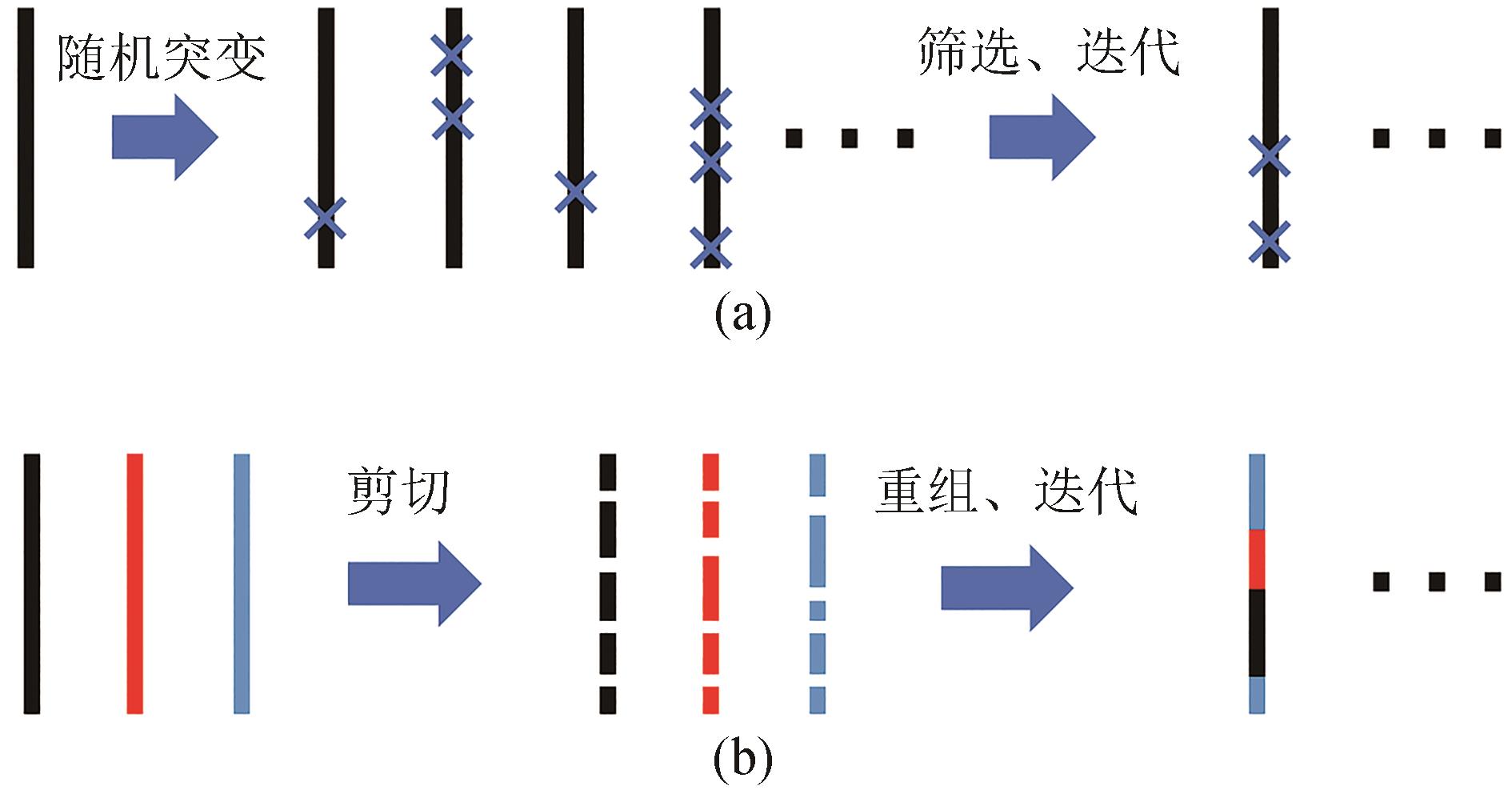
自然界中存在的酶拥有多种多样的功能,它们已经被应用在工业生产和学术研究中,但其中许多酶的性质和功能还不能完全满足应用需要,通过改造来提升这类酶的某些特性是酶工程的重要任务。本文介绍了酶工程的主要发展历程,并重点梳理了人工智能(AI)助力酶工程领域的研究进展。酶工程主要包括理性设计、定向进化、半理性设计和人工智能辅助设计等策略。理性设计方法根据酶的催化机理、结构等先验知识进行改造。定向进化技术通过构建随机突变文库和高通量筛选提升目标酶的稳定性和活性等性质。半理性设计方法借助一系列计算方法构建相比于定向进化更小也更合理的突变文库以降低筛选工作量。人工智能技术在大量数据驱动下可以学习有关蛋白质构成和进化的特征信息。通过直接学习自然界中存在的蛋白质序列、共进化信息和结构,深度神经网络已经可以解决许多类型的酶工程问题,如预测具有有益影响的突变、优化蛋白质的稳定性、提高催化活性等。通过对酶工程现状进行分析,本文旨在进一步推动酶的开发和优化以实现更广泛的应用,为研究者和相关从业人员提供更多有价值的见解。
中图分类号:
引用本文
康里奇, 谈攀, 洪亮. 人工智能时代下的酶工程[J]. 合成生物学, 2023, 4(3): 524-534.
KANG Liqi, TAN Pan, HONG Liang. Enzyme engineering in the age of artificial intelligence[J]. Synthetic Biology Journal, 2023, 4(3): 524-534.
| 蛋白质适应度分类 | 数据集 | ESM-IF1 | ESM-1v | MSA transformer | ProGen2 | Tranception |
|---|---|---|---|---|---|---|
| 催化活性 | B3VI55_LIPST | 0.291 | 0.272 | 0.316 | 0.239 | 0.290 |
| MTH3_HAEAESTABILIZED | 0.423 | 0.488 | 0.564 | 0.507 | 0.479 | |
| KKA2_KLEPN | 0.204 | 0.198 | 0.153 | 0.191 | 0.077 | |
| MK01_HUMAN | 0.155 | 0.164 | 0.182 | 0.200 | 0.203 | |
| AMIE_PSEAE | 0.295 | 0.537 | 0.523 | 0.526 | 0.517 | |
| RASH_HUMAN | 0.070 | 0.131 | 0.089 | 0.135 | 0.085 | |
| UBC9_HUMAN | 0.485 | 0.518 | 0.425 | 0.484 | 0.476 | |
| BG_STRSQ | 0.665 | 0.670 | 0.727 | 0.749 | 0.656 | |
| TRPC_THEMA | 0.392 | 0.488 | 0.462 | 0.397 | 0.444 | |
| TIM_SULSO | 0.506 | 0.617 | 0.613 | 0.529 | 0.594 | |
| P84126_THETH | 0.519 | 0.564 | 0.656 | 0.558 | 0.548 | |
| BLAT_ECOLX | 0.673 | 0.692 | 0.538 | 0.601 | 0.622 | |
| 稳定性 | PTEN_HUMAN | 0.559 | 0.458 | 0.366 | 0.471 | 0.430 |
| TPMT_HUMAN | 0.560 | 0.531 | 0.530 | 0.458 | 0.478 | |
| 肽段结合能力 | DLG4_RAT | 0.468 | 0.531 | 0.224 | 0.361 | 0.446 |
| WW | 0.415 | 0.399 | 0.441 | 0.309 | 0.563 | |
| 蛋白质结合能力 | IF1_ECOLI | 0.337 | 0.356 | 0.363 | 0.246 | 0.347 |
| SUMO1_HUMAN | 0.543 | 0.548 | 0.565 | 0.482 | 0.423 | |
| RL40B_YEAST | 0.372 | 0.365 | 0.647 | 0.473 | 0.455 | |
| DNA结合能力 | FOSJUN | 0.532 | 0.464 | 0.366 | 0.515 | 0.536 |
| GAL4_YEAST | 0.326 | 0.476 | 0.386 | 0.468 | 0.458 | |
| RNA 结合能力 | RRM | 0.443 | 0.536 | 0.509 | 0.512 | 0.407 |
| TDP43 | 0.158 | 0.026 | 0.117 | 0.013 | 0.125 | |
| Ig-G结合能力 | GB1 | 0.337 | 0.105 | 0.329 | 0.232 | 0.254 |
| 平均值 | 0.413 | 0.453 | 0.438 | 0.376 | 0.374 |
表1 无监督模型在不同数据集上预测结果与实验结果的相关性
Table 1 Spearman correlation for predicted fitness developed with unsupervised models
| 蛋白质适应度分类 | 数据集 | ESM-IF1 | ESM-1v | MSA transformer | ProGen2 | Tranception |
|---|---|---|---|---|---|---|
| 催化活性 | B3VI55_LIPST | 0.291 | 0.272 | 0.316 | 0.239 | 0.290 |
| MTH3_HAEAESTABILIZED | 0.423 | 0.488 | 0.564 | 0.507 | 0.479 | |
| KKA2_KLEPN | 0.204 | 0.198 | 0.153 | 0.191 | 0.077 | |
| MK01_HUMAN | 0.155 | 0.164 | 0.182 | 0.200 | 0.203 | |
| AMIE_PSEAE | 0.295 | 0.537 | 0.523 | 0.526 | 0.517 | |
| RASH_HUMAN | 0.070 | 0.131 | 0.089 | 0.135 | 0.085 | |
| UBC9_HUMAN | 0.485 | 0.518 | 0.425 | 0.484 | 0.476 | |
| BG_STRSQ | 0.665 | 0.670 | 0.727 | 0.749 | 0.656 | |
| TRPC_THEMA | 0.392 | 0.488 | 0.462 | 0.397 | 0.444 | |
| TIM_SULSO | 0.506 | 0.617 | 0.613 | 0.529 | 0.594 | |
| P84126_THETH | 0.519 | 0.564 | 0.656 | 0.558 | 0.548 | |
| BLAT_ECOLX | 0.673 | 0.692 | 0.538 | 0.601 | 0.622 | |
| 稳定性 | PTEN_HUMAN | 0.559 | 0.458 | 0.366 | 0.471 | 0.430 |
| TPMT_HUMAN | 0.560 | 0.531 | 0.530 | 0.458 | 0.478 | |
| 肽段结合能力 | DLG4_RAT | 0.468 | 0.531 | 0.224 | 0.361 | 0.446 |
| WW | 0.415 | 0.399 | 0.441 | 0.309 | 0.563 | |
| 蛋白质结合能力 | IF1_ECOLI | 0.337 | 0.356 | 0.363 | 0.246 | 0.347 |
| SUMO1_HUMAN | 0.543 | 0.548 | 0.565 | 0.482 | 0.423 | |
| RL40B_YEAST | 0.372 | 0.365 | 0.647 | 0.473 | 0.455 | |
| DNA结合能力 | FOSJUN | 0.532 | 0.464 | 0.366 | 0.515 | 0.536 |
| GAL4_YEAST | 0.326 | 0.476 | 0.386 | 0.468 | 0.458 | |
| RNA 结合能力 | RRM | 0.443 | 0.536 | 0.509 | 0.512 | 0.407 |
| TDP43 | 0.158 | 0.026 | 0.117 | 0.013 | 0.125 | |
| Ig-G结合能力 | GB1 | 0.337 | 0.105 | 0.329 | 0.232 | 0.254 |
| 平均值 | 0.413 | 0.453 | 0.438 | 0.376 | 0.374 |
| 1 | COBB R E, CHAO R, ZHAO H M. Directed evolution: past, present, and future[J]. AIChE Journal, 2013, 59(5): 1432-1440. |
| 2 | LERNER S A, WU T T, LIN E C. Evolution of a catabolic pathway in bacteria[J]. Science, 1964, 146(3649): 1313-1315. |
| 3 | SARAC I, HOLLENSTEIN M. Terminal deoxynucleotidyl transferase in the synthesis and modification of nucleic acids[J]. ChemBioChem, 2019, 20(7): 860-871. |
| 4 | TOBIN M B, GUSTAFSSON C, HUISMAN G W. Directed evolution: the 'rational' basis for 'irrational' design[J]. Current Opinion in Structural Biology, 2000, 10(4): 421-427. |
| 5 | CHEN K, ARNOLD F H. Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide[J]. Proceedings of the National Academy of Sciences of the United States of America, 1993, 90(12): 5618-5622. |
| 6 | STEMMER W P C. Rapid evolution of a protein in vitro by DNA shuffling[J]. Nature, 1994, 370(6488): 389-391. |
| 7 | STEMMER W P. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution[J]. Proceedings of the National Academy of Sciences of the United States of America, 1994, 91(22): 10747-10751. |
| 8 | LIEBETON K, ZONTA A, SCHIMOSSEK K, et al. Directed evolution of an enantioselective lipase[J]. Chemistry & Biology, 2000, 7(9): 709-718. |
| 9 | REETZ M T, ZONTA A, SCHIMOSSEK K, et al. Creation of enantioselective biocatalysts for organic chemistry by in vitro evolution[J]. Angewandte Chemie International Edition, 1997, 36(24): 2830-2832. |
| 10 | POREBSKI B T, BUCKLE A M. Consensus protein design[J]. Protein Engineering, Design and Selection, 2016, 29(7): 245-251. |
| 11 | STERNKE M, TRIPP K W, BARRICK D. Consensus sequence design as a general strategy to create hyperstable, biologically active proteins[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(23): 11275-11284. |
| 12 | PALMER B, ANGUS K, TAYLOR L, et al. Design of stability at extreme alkaline pH in streptococcal protein G[J]. Journal of Biotechnology, 2008, 134(3/4): 222-230. |
| 13 | MINAKUCHI K, MURATA D, OKUBO Y, et al. Remarkable alkaline stability of an engineered protein A as immunoglobulin affinity ligand: C domain having only one amino acid substitution[J]. Protein Science, 2013, 22(9): 1230-1238. |
| 14 | ROMERO-RIVERA A, GARCIA-BORRÀS M, OSUNA S. Computational tools for the evaluation of laboratory-engineered biocatalysts[J]. Chemical Communications, 2017, 53(2): 284-297. |
| 15 | KHERSONSKY O, LIPSH R, AVIZEMER Z, et al. Automated design of efficient and functionally diverse enzyme repertoires[J]. Molecular Cell, 2018, 72(1): 178-186.e5. |
| 16 | WEINREICH D M, DELANEY N F, DEPRISTO M A, et al. Darwinian evolution can follow only very few mutational paths to fitter proteins[J]. Science, 2006, 312(5770): 111-114. |
| 17 | LI R F, WIJMA H J, SONG L, et al. Computational redesign of enzymes for regio- and enantioselective hydroamination[J]. Nature Chemical Biology, 2018, 14(7): 664-670. |
| 18 | CUI Y L, WANG Y H, TIAN W Y, et al. Development of a versatile and efficient C-N lyase platform for asymmetric hydroamination via computational enzyme redesign[J]. Nature Catalysis, 2021, 4(5): 364-373. |
| 19 | CAPRIOTTI E, FARISELLI P, CASADIO R. A neural-network-based method for predicting protein stability changes upon single point mutations[J]. Bioinformatics, 2004, 20(S1): i63-i68. |
| 20 | CAPRIOTTI E, FARISELLI P, CASADIO R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure[J]. Nucleic Acids Research, 2005, 33(S2): W306-W310. |
| 21 | 曲玉辰, 陆路, 姜世勃. 利用I-Mutant 2.0辅助设计与优化中东呼吸综合征冠状病毒融合抑制多肽[J]. 微生物与感染, 2019, 14(2): 72-81. |
| QU Y C, LU L, JIANG S B. Using I-Mutant2.0 to assist the design and optimization of MERS-CoV fusion inhibitory peptides[J]. Journal of Microbes and Infections, 2019, 14(2): 72-81. | |
| 22 | YANG Y, DING X S, ZHU G C, et al. ProTstab - predictor for cellular protein stability[J]. BMC Genomics, 2019, 20(1): 804. |
| 23 | FARISELLI P, MARTELLI P L, SAVOJARDO C, et al. INPS: predicting the impact of non-synonymous variations on protein stability from sequence[J]. Bioinformatics, 2015, 31(17): 2816-2821. |
| 24 | LAIMER J, HOFER H, FRITZ M, et al. MAESTRO—multi agent stability prediction upon point mutations[J]. BMC Bioinformatics, 2015, 16: 116. |
| 25 | DEHOUCK Y, KWASIGROCH J M, GILIS D, et al. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality[J]. BMC Bioinformatics, 2011, 12: 151. |
| 26 | PIRES D E V, ASCHER D B, BLUNDELL T L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach[J]. Nucleic Acids Research, 2014, 42(W1): W314-W319. |
| 27 | WORTH C L, PREISSNER R, BLUNDELL T L. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic acids research, 2011, 39(S2): W215-W222. |
| 28 | PIRES D E V, ASCHER D B, BLUNDELL T L. mCSM: predicting the effects of mutations in proteins using graph-based signatures[J]. Bioinformatics, 2014, 30(3): 335-342. |
| 29 | The UniProt Consortium. UniProt: the universal protein knowledgebase in 2023[J]. Nucleic Acids Research, 2023, 51(D1): D523-D531. |
| 30 | MEIER J, RAO R S, VERKUIL R, et al. Language models enable zero-shot prediction of the effects of mutations on protein function[C/OL]// Advances in Neural Information Processing Systems 34 (NeurIPS 2021), 2021. 34: 29287-29303 [2023-01-03]. . |
| 31 | RAO R M, LIU J, VERKUIL R, et al. MSA transformer[C/OL]// Proceedings of the 38th International Conference on Machine Learning, PMLR,2021, 139:8844-8856 [2023-01-03]. . |
| 32 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 33 | HSU C, VERKUIL R, LIU J, et al. Learning inverse folding from millions of predicted structures[EB/OL]. bioRxiv, 2022[2023-01-03]. . |
| 34 | JING B, EISMANN S, SURIANA P, et al. Learning from protein structure with geometric vector perceptrons[EB/OL]. arXiv, 2020: 2009.01411[2023-01-03]. . |
| 35 | ZHOU B X, LV O T Y, YI K, et al. Lightweight equivariant graph representation learning for protein engineering[C/OL]//Machine Learning for Structural Biology Workshop - NeurIPS 2022[2023-01-03]. . |
| 36 | RIESSELMAN A J, INGRAHAM J B, MARKS D S. Deep generative models of genetic variation capture the effects of mutations[J]. Nature Methods, 2018, 15(10): 816-822. |
| 37 | NIJKAMP E, RUFFOLO J, WEINSTEIN E N, et al. ProGen2: exploring the boundaries of protein language models[EB/OL]. arXiv, 2022: 2206.13517[2023-01-03]. . |
| 38 | NOTIN P, DIAS M, FRAZER J, et al. Tranception: protein fitness prediction with autoregressive transformers and inference-time retrieval[C/OL]// International Conference on Machine Learning, arXiv, 2022[2023-01-03]. . |
| 39 | LU H Y, DIAZ D J, CZARNECKI N J, et al. Machine learning-aided engineering of hydrolases for PET depolymerization[J]. Nature, 2022, 604(7907): 662-667. |
| 40 | RIVES A, MEIER J, SERCU T, et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(15): e2016239118. |
| 41 | LUO Y N, JIANG G D, YU T H, et al. ECNet is an evolutionary context-integrated deep learning framework for protein engineering[J]. Nature Communications, 2021, 12: 5743. |
| 42 | LI M C, KANG L Q, XIONG Y, et al. SESNet: sequence-structure feature-integrated deep learning method for data-efficient protein engineering[EB/OL]. arXiv, 2022: 2301.00004[2023-01-03]. . |
| 43 | CUI Y L, CHEN Y C, LIU X Y, et al. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy[J]. ACS Catalysis, 2021, 11(3): 1340-1350. |
| 44 | ROCKLIN G J, CHIDYAUSIKU T M, GORESHNIK I, et al. Global analysis of protein folding using massively parallel design, synthesis, and testing[J]. Science, 2017, 357(6347): 168-175. |
| 45 | HUANG B, XU Y, HU X H, et al. A backbone-centred energy function of neural networks for protein design[J]. Nature, 2022, 602(7897): 523-528. |
| 46 | DOU J Y, VOROBIEVA A A, SHEFFLER W, et al. De novo design of a fluorescence-activating β-barrel[J]. Nature, 2018, 561(7724): 485-491. |
| 47 | YEH A H W, NORN C, KIPNIS Y, et al. De novo design of luciferases using deep learning[J]. Nature, 2023, 614(7949): 774-780. |
| 48 | RUSS W P, FIGLIUZZI M, STOCKER C, et al. An evolution-based model for designing chorismate mutase enzymes[J]. Science, 2020, 369(6502): 440-445. |
| 49 | REPECKA D, JAUNISKIS V, KARPUS L, et al. Expanding functional protein sequence spaces using generative adversarial networks[J]. Nature Machine Intelligence, 2021, 3(4): 324-333. |
| 50 | MADANI A, KRAUSE B, GREENE E R, et al. Large language models generate functional protein sequences across diverse families[J/OL]. Nature Biotechnology, 2023[2023-02-01]. . |
| 51 | SINAI S, WANG R, WHATLEY A, et al. AdaLead: a simple and robust adaptive greedy search algorithm for sequence design[EB/OL]. arXiv, 2020: 2010.02141[2023-01-03]. . |
| 52 | BISWAS S, KHIMULYA G, ALLEY E C, et al. Low-N protein engineering with data-efficient deep learning[J]. Nature Methods, 2021, 18(4): 389-396. |
| 53 | HU R Y, FU L H, CHEN Y C, et al. Protein engineering via Bayesian optimization-guided evolutionary algorithm and robotic experiments[J]. Briefings in Bioinformatics, 2023, 24(1): bbac570. |
| 54 | CASTRO E, GODAVARTHI A, RUBINFIEN J, et al. Transformer-based protein generation with regularized latent space optimization[J]. Nature Machine Intelligence, 2022, 4(10): 840-851. |
| 55 | LIN Z, AKIN H, RAO R, et al. Evolutionary-scale prediction of atomic-level protein structure with a language model[J]. Science, 2023, 379(6637): 1123-1130. |
| [1] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [2] | 付雨, 钟芳锐. 化学原理驱动的光生物不对称催化研究进展[J]. 合成生物学, 2024, 5(5): 1021-1049. |
| [3] | 汤志军, 胡友财, 刘文. 酶促4+2和2+2环加成反应:区域与立体选择性的理解与应用[J]. 合成生物学, 2024, 5(3): 401-407. |
| [4] | 张俊, 金诗雪, 云倩, 瞿旭东. 聚酮化合物非天然延伸单元的生物合成与结构改造应用[J]. 合成生物学, 2024, 5(3): 561-570. |
| [5] | 奚萌宇, 胡逸灵, 顾玉诚, 戈惠明. 基因组挖掘指导天然药物分子的发现[J]. 合成生物学, 2024, 5(3): 447-473. |
| [6] | 雷茹, 陶慧, 刘天罡. 基因组深度挖掘驱动微生物萜类化合物高效发现[J]. 合成生物学, 2024, 5(3): 507-526. |
| [7] | 朱景勇, 李钧翔, 李旭辉, 张瑾, 毋文静. 深度学习在基于序列的蛋白质互作预测中的应用进展[J]. 合成生物学, 2024, 5(1): 88-106. |
| [8] | 孙梦楚, 陆亮宇, 申晓林, 孙新晓, 王佳, 袁其朋. 基于荧光检测的高通量筛选技术和装备助力细胞工厂构建[J]. 合成生物学, 2023, 4(5): 947-965. |
| [9] | 明阳, 陈彬, 黄小强. 光酶催化合成进展[J]. 合成生物学, 2023, 4(4): 651-675. |
| [10] | 孟巧珍, 郭菲. “可折叠性”在酶智能设计改造中的应用研究——以AlphaFold2为例[J]. 合成生物学, 2023, 4(3): 571-589. |
| [11] | 陈志航, 季梦麟, 戚逸飞. 人工智能蛋白质结构设计算法研究进展[J]. 合成生物学, 2023, 4(3): 464-487. |
| [12] | 赖奇龙, 姚帅, 查毓国, 白虹, 宁康. 微生物组生物合成基因簇发掘方法及应用前景[J]. 合成生物学, 2023, 4(3): 611-627. |
| [13] | 宋益东, 袁乾沐, 杨跃东. 深度学习在蛋白质功能预测中的应用[J]. 合成生物学, 2023, 4(3): 488-506. |
| [14] | 王晟, 王泽琛, 陈威华, 陈珂, 彭向达, 欧发芬, 郑良振, 孙瑨原, 沈涛, 赵国屏. 基于人工智能和计算生物学的合成生物学元件设计[J]. 合成生物学, 2023, 4(3): 422-443. |
| [15] | 阮青云, 黄莘, 孟子钧, 全舒. 蛋白质稳定性计算设计与定向进化前沿工具[J]. 合成生物学, 2023, 4(1): 5-29. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||