合成生物学 ›› 2025, Vol. 6 ›› Issue (2): 320-333.DOI: 10.12211/2096-8280.2024-071
乳酸菌的合成生物学工具及在合成益肤因子中的应用
郭婷婷, 韩湘凝, 黄熙婷, 张婷婷, 孔健
- 山东大学微生物改造技术全国重点实验室,山东 青岛 266237
-
收稿日期:2024-09-11修回日期:2024-11-14出版日期:2025-04-30发布日期:2025-05-20 -
通讯作者:孔健 -
作者简介:郭婷婷 (1984—),女,副教授,硕士生导师。研究方向为乳酸菌遗传操作系统、乳酸菌合成生物学与活性产物的高效生产。E-mail:guotingting@sdu.edu.cn孔健 (1964—),女,教授,博士生导师,研究方向为乳酸菌生理遗传及其应用,乳酸菌生物活性物质的开发等。E-mail:kongjian@sdu.edu.cn -
基金资助:微生物改造技术全国重点实验室“揭榜挂帅”项目(SKLMTFCP-2023-05)
Advances in synthetic biology tools for lactic acid bacteria and their application in the development of skin beneficial products
GUO Tingting, HAN Xiangning, HUANG Xiting, ZHANG Tingting, KONG Jian
- State Key Laboratory of Microbial Technology,Shandong University,Qingdao 266237,Shandong,China
-
Received:2024-09-11Revised:2024-11-14Online:2025-04-30Published:2025-05-20 -
Contact:KONG Jian
摘要:
乳酸菌在发酵食品中的使用有着悠久历史,一些菌种被认为是安全级微生物。乳酸菌也是人体正常的共生菌,具有调节肠道和皮肤微生态的菌群平衡、增强机体免疫力等有益作用。当前,乳酸菌及其产生的活性代谢产物添加到护肤品中产生保湿、抗氧化和减轻过敏等效果得到了消费者认可,而一些乳酸菌在缓解和治疗皮肤疾病方面的作用也有越来越多的数据支持。基于此,利用乳酸菌作为底盘生产护肤活性物质或作为皮肤修复的生物治疗载体具有广阔的应用前景。本文首先总结了在乳酸菌中建立的遗传操作系统,着重关注遗传可及性、基因表达系统和基因组编辑技术,然后总结了乳酸菌作为细胞工厂生产保湿因子和抗氧化产物的研究进展,最后介绍了工程乳酸菌针对皮肤损伤进行靶向递药的可行性,旨在为乳酸菌应用于皮肤健康领域提供借鉴。
中图分类号:
引用本文
郭婷婷, 韩湘凝, 黄熙婷, 张婷婷, 孔健. 乳酸菌的合成生物学工具及在合成益肤因子中的应用[J]. 合成生物学, 2025, 6(2): 320-333.
GUO Tingting, HAN Xiangning, HUANG Xiting, ZHANG Tingting, KONG Jian. Advances in synthetic biology tools for lactic acid bacteria and their application in the development of skin beneficial products[J]. Synthetic Biology Journal, 2025, 6(2): 320-333.
| 1 | GRICE E A, SEGRE J A. The skin microbiome[J]. Nature Reviews Microbiology, 2011, 9(4): 244-253. |
| 2 | HARRIS-TRYON T A, GRICE E A. Microbiota and maintenance of skin barrier function[J]. Science, 2022, 376(6596): 940-945. |
| 3 | GEOGHEGAN J A, IRVINE A D, FOSTER T J. Staphylococcus aureus and atopic dermatitis: a complex and evolving relationship[J]. Trends in Microbiology, 2018, 26(6): 484-497. |
| 4 | BRANDWEIN M, FUKS G, ISRAEL A, et al. Skin microbiome compositional changes in atopic dermatitis accompany dead sea climatotherapy[J]. Photochemistry and Photobiology, 2019, 95(6): 1446-1453. |
| 5 | CHEN Y H, SONG Y P, CHEN Z G, et al. Early-life skin microbial biomarkers for eczema phenotypes in Chinese toddlers[J]. Pathogens, 2023, 12(5): 697. |
| 6 | WANG H L, CHAN M W M, CHAN H H, et al. Longitudinal changes in skin microbiome associated with change in skin status in patients with psoriasis[J]. Acta Dermato-Venereologica, 2020, 100(18): adv00329. |
| 7 | PLAVEC T V, BERLEC A. Safety aspects of genetically modified lactic acid bacteria[J]. Microorganisms, 2020, 8(2): 297. |
| 8 | MASOOD M I, QADIR M I, SHIRAZI J H, et al. Beneficial effects of lactic acid bacteria on human beings[J]. Critical Reviews in Microbiology, 2011, 37(1): 91-98. |
| 9 | DOU J X, FENG N, GUO F Y, et al. Applications of probiotic constituents in cosmetics[J]. Molecules, 2023, 28(19): 6765. |
| 10 | LEBEER S, OERLEMANS E F M, CLAES I, et al. Selective targeting of skin pathobionts and inflammation with topically applied lactobacilli[J]. Cell Reports Medicine, 2022, 3(2): 100521. |
| 11 | MOTTIN BPHARM V H M, SUYENAGA E S. An approach on the potential use of probiotics in the treatment of skin conditions: acne and atopic dermatitis[J]. International Journal of Dermatology, 2018, 57(12): 1425-1432. |
| 12 | CHRISTENSEN I B, VEDEL C, CLAUSEN M L, et al. Targeted screening of lactic acid bacteria with antibacterial activity toward Staphylococcus aureus clonal complex type 1 associated with atopic dermatitis[J]. Frontiers in Microbiology, 2021, 12: 733847. |
| 13 | ALSAHEB R A ABD, ALADDIN A, OTHMAN N Z, et al. Lactic acid applications in pharmaceutical and cosmeceutical industries[J]. Journal of Chemical and Pharmaceutical Research, 2015, 7(10): 729-735. |
| 14 | BOLOTIN A, WINCKER P, MAUGER S, et al. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403[J]. Genome Research, 2001, 11(5): 731-753. |
| 15 | KOK J, VAN GIJTENBEEK L A, DE JONG A, et al. The evolution of gene regulation research in Lactococcus lactis [J]. FEMS Microbiology Reviews, 2017, 41(Supp_1): S220-S243. |
| 16 | PEDERSEN M B, GARRIGUES C, TUPHILE K, et al. Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon[J]. Journal of Bacteriology, 2008, 190(14): 4903-4911. |
| 17 | LIU J M, WANG Z H, KANDASAMY V, et al. Harnessing the respiration machinery for high-yield production of chemicals in metabolically engineered Lactococcus lactis [J]. Metabolic Engineering, 2017, 44: 22-29. |
| 18 | NEEF J, KOEDIJK D G A M, BOSMA T, et al. Efficient production of secreted staphylococcal antigens in a non-lysing and proteolytically reduced Lactococcus lactis strain[J]. Applied Microbiology and Biotechnology, 2014, 98(24): 10131-10141. |
| 19 | CAMPOS G M, AMÉRICO M F, DOS SANTOS FREITAS A, et al. Lactococcus lactis as an interleukin delivery system for prophylaxis and treatment of inflammatory and autoimmune diseases[J]. Probiotics and Antimicrobial Proteins, 2024, 16(2): 352-366. |
| 20 | URIOT O, DENIS S, JUNJUA M, et al. Streptococcus thermophilus: from yogurt starter to a new promising probiotic candidate?[J]. Journal of Functional Foods, 2017, 37: 74-89. |
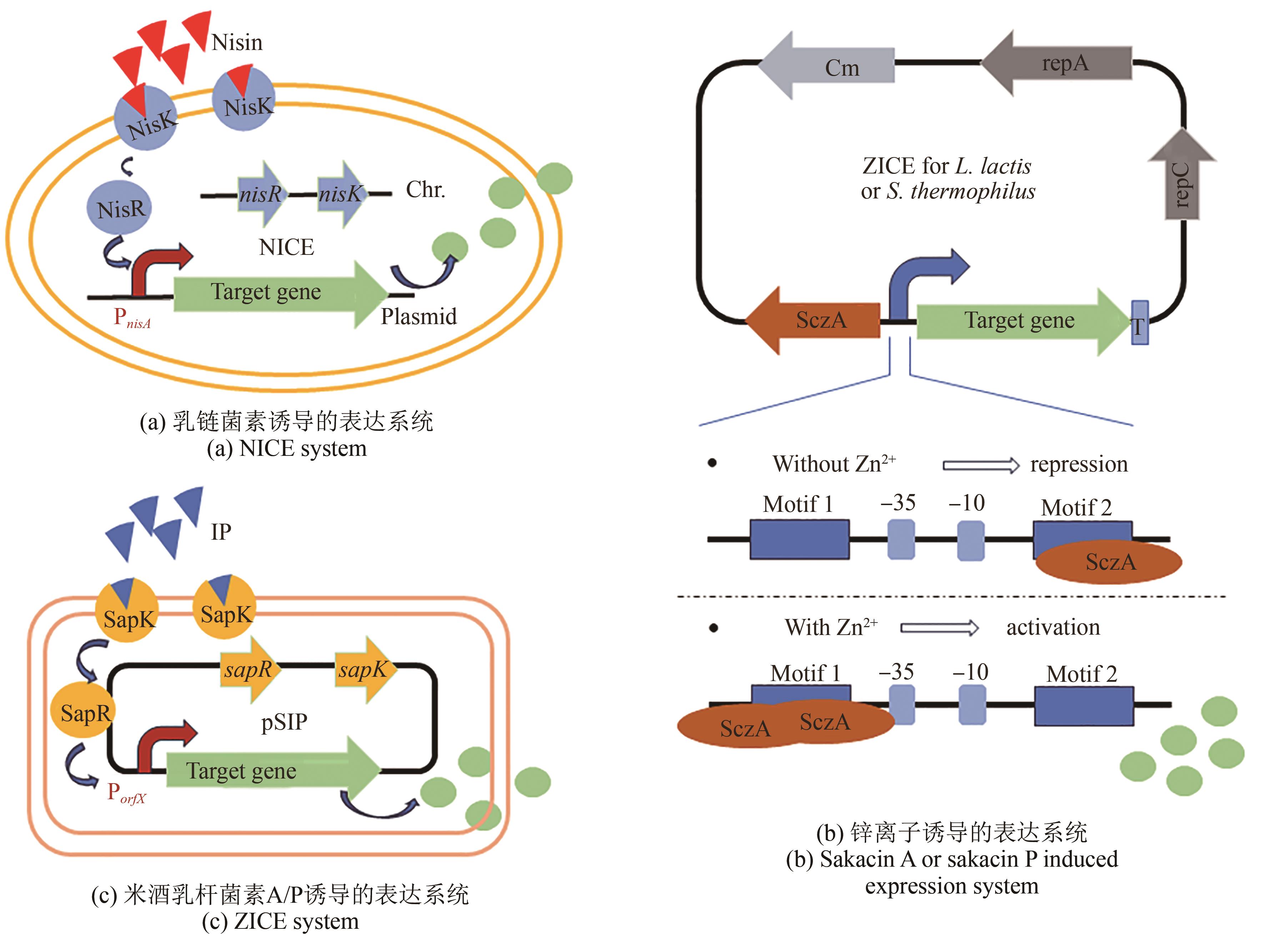
| 21 | MARKAKIOU S, GASPAR P, JOHANSEN E, et al. Harnessing the metabolic potential of Streptococcus thermophilus for new biotechnological applications[J]. Current Opinion in Biotechnology, 2020, 61: 142-152. |
| 22 | SUN Z H, HARRIS H M B, MCCANN A, et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated Genera[J]. Nature Communications, 2015, 6: 8322. |
| 23 | XIN Y P, MU Y L, KONG J, et al. Targeted and repetitive chromosomal integration enables high-level heterologous gene expression in Lactobacillus casei [J]. Applied and Environmental Microbiology, 2019, 85(9): e00033-19. |
| 24 | ZHOU D, JIANG Z N, PANG Q X, et al. CRISPR/Cas9-assisted seamless genome editing in Lactobacillus plantarum and its application in N-acetylglucosamine production[J]. Applied and Environmental Microbiology, 2019, 85(21): e01367-19. |
| 25 | BRON P A, MARCELLI B, MULDER J, et al. Renaissance of traditional DNA transfer strategies for improvement of industrial lactic acid bacteria[J]. Current Opinion in Biotechnology, 2019, 56: 61-68. |
| 26 | AMMANN A, NEVE H, GEIS A, et al. Plasmid transfer via transduction from Streptococcus thermophilus to Lactococcus lactis [J]. Journal of Bacteriology, 2008, 190(8): 3083-3087. |
| 27 | LAMPKOWSKA J, FELD L, MONAGHAN A, et al. A standardized conjugation protocol to asses antibiotic resistance transfer between lactococcal species[J]. International Journal of Food Microbiology, 2008, 127(1-2): 172-175. |
| 28 | DANDOY D, FREMAUX C, DE FRAHAN M H, et al. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS[J]. Microbial Cell Factories, 2011, 10(): S21. |
| 29 | MULDER J, WELS M, KUIPERS O P, et al. Unleashing natural competence in Lactococcus lactis by induction of the competence regulator ComX[J]. Applied and Environmental Microbiology, 2017, 83(20): e01320-17. |
| 30 | DAVID B, RADZIEJWOSKI A, TOUSSAINT F, et al. Natural DNA transformation is functional in Lactococcus lactis subsp. cremoris KW2[J]. Applied and Environmental Microbiology, 2017, 83(16): e01074-17. |
| 31 | MORAWSKA L P, KUIPERS O P. Cell-to-cell non-conjugative plasmid transfer between Bacillus subtilis and lactic acid bacteria[J]. Microbial Biotechnology, 2023, 16(4): 784-798. |
| 32 | HOLO H, NES I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media[J]. Applied and Environmental Microbiology, 1989, 55(12): 3119-3123. |
| 33 | NATORI Y, KANO Y, IMAMOTO F. Genetic transformation of Lactobacillus casei by electroporation[J]. Biochimie, 1990, 72(4): 265-269. |
| 34 | KONG L H, XIONG Z Q, XIA Y J, et al. High-efficiency transformation of Streptococcus thermophilus using electroporation[J]. Journal of the Science of Food and Agriculture, 2021, 101(15): 6578-6585. |
| 35 | MENG Q, YUAN Y X, LI Y Y, et al. Optimization of electrotransformation parameters and engineered promoters for Lactobacillus plantarum from wine[J]. ACS Synthetic Biology, 2021, 10(7): 1728-1738. |
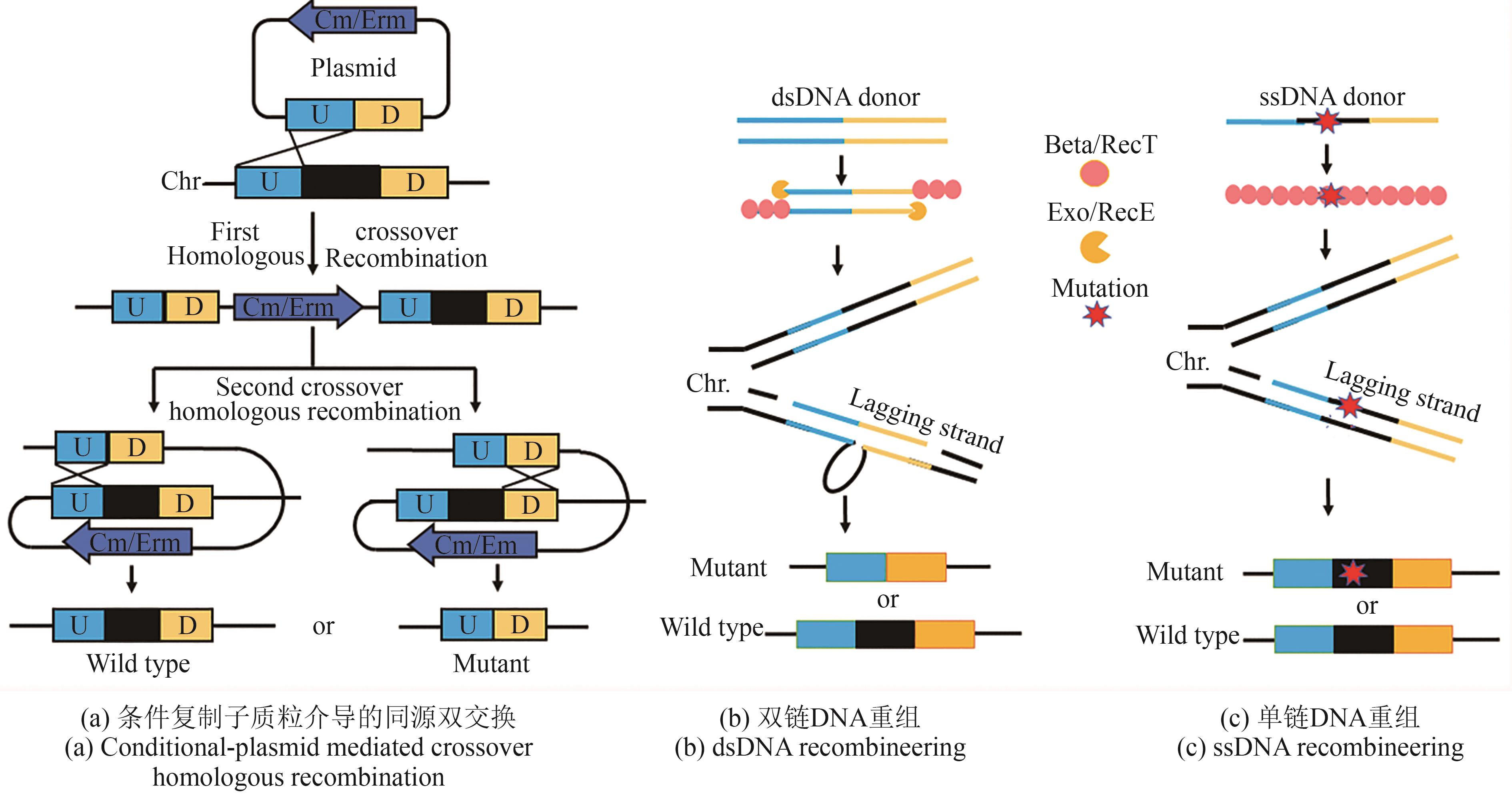
| 36 | 陈韫慧, 夏永军, 宋馨, 等. 乳酸菌作为生物活性物质体内递送载体的研究进展[J]. 食品科学, 2023, 44(13): 193-202. |
| CHEN Y H, XIA Y J, SONG X, et al. Advances in the study of lactic acid bacteria as vehicles for the in vivo delivery of bioactive substances[J]. Food Science, 2023, 44(13): 193-202. | |
| 37 | ZHU D L, LIU F L, XU H J, et al. Isolation of strong constitutive promoters from Lactococcus lactis subsp. lactis N8[J]. FEMS Microbiology Letters, 2015, 362(16): fnv107. |
| 38 | NARITA J, ISHIDA S, OKANO K, et al. Improvement of protein production in lactic acid bacteria using 5′-untranslated leader sequence of slpA from Lactobacillus acidophilus. Improvement in protein production using UTLS[J]. Applied Microbiology and Biotechnology, 2006, 73(2): 366-373. |
| 39 | SUN W H, JIANG B, ZHANG Y, et al. Enabling the biosynthesis of malic acid in Lactococcus lactis by establishing the reductive TCA pathway and promoter engineering[J]. Biochemical Engineering Journal, 2020, 161: 107645. |
| 40 | PEIROTÉN Á, LANDETE J M. Natural and engineered promoters for gene expression in Lactobacillus species[J]. Applied Microbiology and Biotechnology, 2020, 104(9): 3797-3805. |
| 41 | KUIPERS O P, BEERTHUYZEN M M, DE RUYTER P G G A, et al. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction[J]. Journal of Biological Chemistry, 1995, 270(45): 27299-27304. |
| 42 | MIERAU I, KLEEREBEZEM M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis [J]. Applied Microbiology and Biotechnology, 2005, 68(6): 705-717. |
| 43 | RENYE J A JR, SOMKUTI G A. Nisin-induced expression of pediocin in dairy lactic acid bacteria[J]. Journal of Applied Microbiology, 2010, 108(6): 2142-2151. |
| 44 | YANG P, WANG J, QI Q S. Prophage recombinases-mediated genome engineering in Lactobacillus plantarum [J]. Microbial Cell Factories, 2015, 14: 154. |
| 45 | LLULL D, POQUET I. New expression system tightly controlled by zinc availability in Lactococcus lactis [J]. Applied and Environmental Microbiology, 2004, 70(9): 5398-5406. |
| 46 | MIYOSHI A, JAMET E, COMMISSAIRE J, et al. A xylose-inducible expression system for Lactococcus lactis [J]. FEMS Microbiology Letters, 2004, 239(2): 205-212. |
| 47 | MADSEN S M, ARNAU J, VRANG A, et al. Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis [J]. Molecular Microbiology, 1999, 32(1): 75-87. |
| 48 | XU X N, ZHANG L W, CUI Y, et al. Development of Zn2+-controlled expression system for lactic acid bacteria and its application in engineered probiotics[J]. Synthetic and Systems Biotechnology, 2024, 9(1): 152-158. |
| 49 | AXELSSON L, LINDSTAD G, NATERSTAD K. Development of an inducible gene expression system for Lactobacillus sakei [J]. Letters in Applied Microbiology, 2003, 37(2): 115-120. |
| 50 | JIMÉNEZ J J, DIEP D B, BORRERO J, et al. Cloning strategies for heterologous expression of the bacteriocin enterocin A by Lactobacillus sakei Lb790, Lb. plantarum NC8 and Lb. casei CECT475[J]. Microbial Cell Factories, 2015, 14: 166. |
| 51 | SØRVIG E, MATHIESEN G, NATERSTAD K, et al. High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors[J]. Microbiology, 2005, 151(Pt 7): 2439-2449. |
| 52 | FILSINGER G T, WANNIER T M, PEDERSEN F B, et al. Characterizing the portability of phage-encoded homologous recombination proteins[J]. Nature Chemical Biology, 2021, 17(4): 394-402. |
| 53 | MARKAKIOU S, NEVES A R, ZEIDAN A A, et al. Development of a tetracycline-inducible system for conditional gene expression in Lactococcus lactis and Streptococcus thermophilus [J]. Microbiology Spectrum, 2023, 11(3): e00668-23. |
| 54 | HEISS S, HÖRMANN A, TAUER C, et al. Evaluation of novel inducible promoter/repressor systems for recombinant protein expression in Lactobacillus plantarum [J]. Microbial Cell Factories, 2016, 15: 50. |
| 55 | BÖRNER R A, KANDASAMY V, AXELSEN A M, et al. Genome editing of lactic acid bacteria: opportunities for food, feed, pharma and biotech[J]. FEMS Microbiology Letters, 2019, 366(1): fny291. |
| 56 | LEENHOUTS K J, KOK J, VENEMA G. Lactococcal plasmid pWV01 as an integration vector for lactococci[J]. Applied and Environmental Microbiology, 1991, 57(9): 2562-2567. |
| 57 | MAGUIN E, DUWAT P, HEGE T, et al. New thermosensitive plasmid for Gram-positive bacteria[J]. Journal of Bacteriology, 1992, 174(17): 5633-5638. |
| 58 | XIN Y P, GUO T T, MU Y L, et al. Development of a counterselectable seamless mutagenesis system in lactic acid bacteria[J]. Microbial Cell Factories, 2017, 16(1): 116. |
| 59 | SOLEM C, DEFOOR E, JENSEN P R, et al. Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis [J]. Applied and Environmental Microbiology, 2008, 74(15): 4772-4775. |
| 60 | GOH Y J, AZCÁRATE-PERIL M A, O’FLAHERTY S, et al. Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM[J]. Applied and Environmental Microbiology, 2009, 75(10): 3093-3105. |
| 61 | PETERSEN K V, MARTINUSSEN J, JENSEN P R, et al. Repetitive, marker-free, site-specific integration as a novel tool for multiple chromosomal integration of DNA[J]. Applied and Environmental Microbiology, 2013, 79(12): 3563-3569. |
| 62 | RAWSTHORNE H, TURNER K N, MILLS D A. Multicopy integration of heterologous genes, using the lactococcal group Ⅱ intron targeted to bacterial insertion sequences[J]. Applied and Environmental Microbiology, 2006, 72(9): 6088-6093. |
| 63 | ZHANG Y, BUCHHOLZ F, MUYRERS J P, et al. A new logic for DNA engineering using recombination in Escherichia coli [J]. Nature Genetics, 1998, 20(2): 123-128. |
| 64 | YU D G, ELLIS H M, LEE E C, et al. An efficient recombination system for chromosome engineering in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(11): 5978-5983. |
| 65 | CHAI Y, SHAN S P, WEISSMAN K J, et al. Heterologous expression and genetic engineering of the tubulysin biosynthetic gene cluster using red/ET recombineering and inactivation mutagenesis[J]. Chemistry & Biology, 2012, 19(3): 361-371. |
| 66 | STRINGER A M, SINGH N, YERMAKOVA A, et al. FRUIT, a scar-free system for targeted chromosomal mutagenesis, epitope tagging, and promoter replacement in Escherichia coli and Salmonella enterica [J]. PLoS One, 2012, 7(9): e44841. |
| 67 | XIN Y P, GUO T T, MU Y L, et al. Identification and functional analysis of potential prophage-derived recombinases for genome editing in Lactobacillus casei [J]. FEMS Microbiology Letters, 2017, 364(24): fnx243. |
| 68 | HUANG H, SONG X, YANG S. Development of a RecE/T-assisted CRAISPR-Cas9 toolbox for Lactobacillus [J]. Biotechnology Journal, 2019, 14(7): 1800690. |
| 69 | VAN PIJKEREN J P, BRITTON R A. High efficiency recombineering in lactic acid bacteria[J]. Nucleic Acids Research, 2012, 40(10): e76. |
| 70 | HAO M Y, CUI Y H, QU X J. Analysis of CRISPR-Cas system in Streptococcus thermophilus and its application[J]. Frontiers in Microbiology, 2018, 9: 257. |
| 71 | YANG L, LI W X, UJIROGHENE O J, et al. Occurrence and diversity of CRISPR loci in Lactobacillus casei group[J]. Frontiers in Microbiology, 2020, 11: 624. |
| 72 | SCHUSTER J A, VOGEL R F, EHRMANN M A. Characterization and distribution of CRISPR-Cas systems in Lactobacillus sakei [J]. Archives of Microbiology, 2019, 201(3): 337-347. |
| 73 | MA S, WANG F Y, ZHANG X J, et al. Repurposing endogenous type Ⅱ CRISPR-Cas9 system for genome editing in Streptococcus thermophilus [J]. Biotechnology and Bioengineering, 2024, 121(2): 749-756. |
| 74 | MARTEL B, MOINEAU S. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages[J]. Nucleic Acids Research, 2014, 42(14): 9504-9513. |
| 75 | SONG X, HUANG H, XIONG Z Q, et al. CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei [J]. Applied and Environmental Microbiology, 2017, 83(22): e01259-17. |
| 76 | OH J H, VAN PIJKEREN J P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri [J]. Nucleic Acids Research, 2014, 42(17): e131. |
| 77 | GUO T T, XIN Y P, ZHANG Y, et al. A rapid and versatile tool for genomic engineering in Lactococcus lactis [J]. Microbial Cell Factories, 2019, 18(1): 22. |
| 78 | KONG L H, SONG X, XIA Y J, et al. Construction of a CRISPR/nCas9-assisted genome editing system for exopolysaccharide biosynthesis in Streptococcus thermophilus [J]. Food Research International, 2022, 158: 111550. |
| 79 | TIAN K R, HONG X, GUO M M, et al. Development of base editors for simultaneously editing multiple loci in Lactococcus lactis [J]. ACS Synthetic Biology, 2022, 11(11): 3644-3656. |
| 80 | VAN TILBURG A Y, CAO H J, VAN DER MEULEN S B, et al. Metabolic engineering and synthetic biology employing Lactococcus lactis and Bacillus subtilis cell factories[J]. Current Opinion in Biotechnology, 2019, 59: 1-7. |
| 81 | SHENG J Z, LING P X, ZHU X Q, et al. Use of induction promoters to regulate hyaluronan synthase and UDP-glucose-6-dehydrogenase of Streptococcus zooepidemicus expression in Lactococcus lactis: a case study of the regulation mechanism of hyaluronic acid polymer[J]. Journal of Applied Microbiology, 2009, 107(1): 136-144. |
| 82 | ZHONG Q, MA Y Q, XU D L, et al. Heterologous biosynthesis of hyaluronic acid using a new hyaluronic acid synthase derived from the probiotic Streptococcus thermophilus [J]. Fermentation, 2023, 9(6): 510. |
| 83 | MOHAMMED A A, NIAMAH A K. Identification and antioxidant activity of hyaluronic acid extracted from local isolates of Streptococcus thermophilus [J]. Materials Today: Proceedings, 2022, 60: 1523-1529. |
| 84 | LEW L C, GAN C Y, LIONG M T. Dermal bioactives from lactobacilli and bifidobacteria[J]. Annals of Microbiology, 2013, 63(3): 1047-1055. |
| 85 | SUNGUROĞLU C, SEZGIN D E, ÇELIK P A, et al. Higher titer hyaluronic acid production in recombinant Lactococcus lactis [J]. Preparative Biochemistry & Biotechnology, 2018, 48(8): 734-742. |
| 86 | PRASAD S B, JAYARAMAN G, RAMACHANDRAN K B. Hyaluronic acid production is enhanced by the additional co-expression of UDP-glucose pyrophosphorylase in Lactococcus lactis [J]. Applied Microbiology and Biotechnology, 2010, 86(1): 273-283. |
| 87 | HMAR R V, PRASAD S B, JAYARAMAN G, et al. Chromosomal integration of hyaluronic acid synthesis (has) genes enhances the molecular weight of hyaluronan produced in Lactococcus lactis [J]. Biotechnology Journal, 2014, 9(12): 1554-1564. |
| 88 | JEEVA P, SHANMUGA DOSS S, SUNDARAM V, et al. Production of controlled molecular weight hyaluronic acid by glucostat strategy using recombinant Lactococcus lactis cultures[J]. Applied Microbiology and Biotechnology, 2019, 103(11): 4363-4375. |
| 89 | 张少伦, 高聪, 李晓敏, 等. 代谢工程改造克雷伯氏菌生产1,3-丙二醇[J]. 生物工程学报, 2024, 40(8): 2386-2402. |
| ZHANG S L, GAO C, LI X M, et al. Metabolic engineering of Klebsiella pneumoniae for 1,3-propanediol production[J]. Chinese Journal of Biotechnology, 2024, 40(8): 2386-2402. | |
| 90 | 刘建明, 张炽坚, 张冰, 等. 巴氏梭菌作为工业底盘细胞高效生产1,3-丙二醇——从代谢工程和菌种进化到过程工程和产品分离[J]. 合成生物学, 2024, 5(6): 1386-1403. |
| LIU J M, ZHANG Z J, ZHANG B, et al. Clostridium pasteurianum as an industrial chassis for efficient productionof 1,3-propanediol: from metabolic engineering to fermentation andproduct separation[J]. Synthetic Biology Journal, 2024, 5(6): 1386-1403. | |
| 91 | 蒋欢, 马江山, 曾柏全, 等. 粗甘油发酵生产1,3-丙二醇的研究进展[J]. 生物技术通报, 2022, 38(10): 45-53. |
| JIANG H, MA J S, ZENG B Q, et al. Research progress in 1,3-propanediol production by fermenting crude glycerol[J]. Biotechnology Bulletin, 2022, 38(10): 45-53. | |
| 92 | WU Y X, LIN Y H. Fermentation redox potential control on the 1,3-propanediol production by Lactobacillus panis PM1[J]. Process Biochemistry, 2022, 114: 139-146. |
| 93 | JU J H, WANG D X, HEO S Y, et al. Enhancement of 1,3-propanediol production from industrial by-product by Lactobacillus reuteri CH53[J]. Microbial Cell Factories, 2020, 19(1): 6. |
| 94 | JU J H, HEO S Y, CHOI S W, et al. Effective bioconversion of 1,3-propanediol from biodiesel-derived crude glycerol using organic acid resistance-enhanced Lactobacillus reuteri JH83[J]. Bioresource Technology, 2021, 337: 125361. |
| 95 | SINGH K, AINALA S K, PARK S. Metabolic engineering of Lactobacillus reuteri DSM 20, 016 for improved 1,3-propanediol production from glycerol[J]. Bioresource Technology, 2021, 338: 125590. |
| 96 | SINGH K, PARK S. Construction of prophage-free and highly-transformable Limosilactobacillus reuteri strains and their use for production of 1,3-propanediol[J]. Biotechnology and Bioengineering, 2024, 121(1): 317-328. |
| 97 | HAWKINS C L, DAVIES M J. Detection, identification, and quantification of oxidative protein modifications[J]. Journal of Biological Chemistry, 2019, 294(51): 19683-19708. |
| 98 | KONG L H, XIONG Z Q, SONG X, et al. Characterization of a panel of strong constitutive promoters from Streptococcus thermophilus for fine-tuning gene expression[J]. ACS Synthetic Biology, 2019, 8(6): 1469-1472. |
| 99 | LIN J Z, ZOU Y X, CAO K L, et al. The impact of heterologous catalase expression and superoxide dismutase overexpression on enhancing the oxidative resistance in Lactobacillus casei [J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(5): 703-711. |
| 100 | AN H R, ZHAI Z Y, YIN S, et al. Coexpression of the superoxide dismutase and the catalase provides remarkable oxidative stress resistance in Lactobacillus rhamnosus [J]. Journal of Agricultural and Food Chemistry, 2011, 59(8): 3851-3856. |
| 101 | LI Y, HUGENHOLTZ J, SYBESMA W, et al. Using Lactococcus lactis for glutathione overproduction[J]. Applied Microbiology and Biotechnology, 2005, 67(1): 83-90. |
| 102 | XU C T, SHI Z W, SHAO J Q, et al. Metabolic engineering of Lactococcus lactis for high level accumulation of glutathione and S-adenosyl-L-methionine[J]. World Journal of Microbiology & Biotechnology, 2019, 35(12): 185. |
| 103 | WU J P, TIAN X F, XU X N, et al. Engineered probiotic Lactococcus lactis for lycopene production against ROS stress in intestinal epithelial cells[J]. ACS Synthetic Biology, 2022, 11(4): 1568-1576. |
| 104 | MA J, LI C Y, WANG J R, et al. Genetically engineered Escherichia coli nissle 1917 secreting GLP-1 analog exhibits potential antiobesity effect in high-fat diet-induced obesity mice[J]. Obesity, 2020, 28(2): 315-322. |
| 105 | CUBILLOS-RUIZ A, ALCANTAR M A, DONGHIA N M, et al. An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis[J]. Nature Biomedical Engineering, 2022, 6(7): 910-921. |
| 106 | SCOTT B M, GUTIÉRREZ-VÁZQUEZ C, SANMARCO L M, et al. Self-tunable engineered yeast probiotics for the treatment of inflammatory bowel disease[J]. Nature Medicine, 2021, 27(7): 1212-1222. |
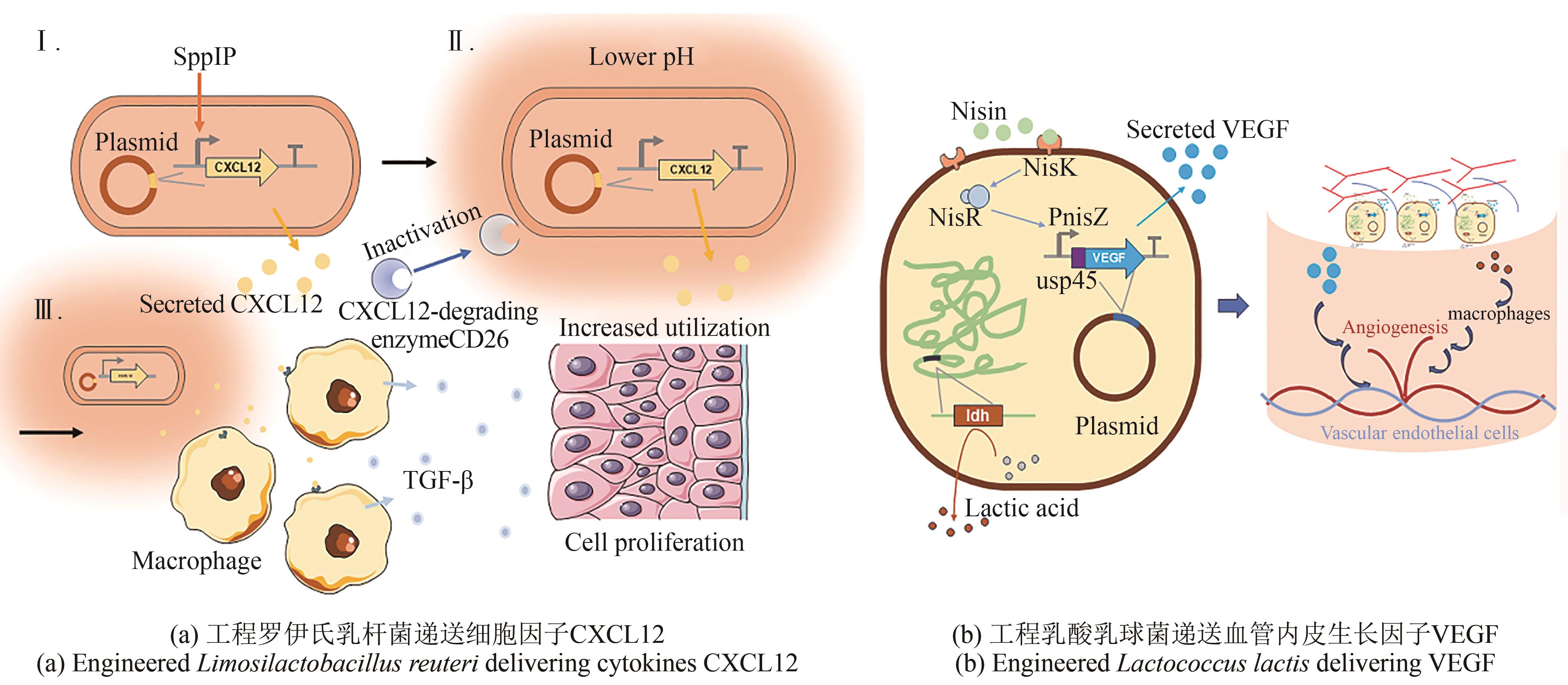
| 107 | VÅGESJÖ E, ÖHNSTEDT E, MORTIER A, et al. Accelerated wound healing in mice by on-site production and delivery of CXCL12 by transformed lactic acid bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(8): 1895-1900. |
| 108 | ÖHNSTEDT E, LOFTON TOMENIUS H, FRANK P, et al. Accelerated wound healing in minipigs by on-site production and delivery of CXCL12 by transformed lactic acid bacteria[J]. Pharmaceutics, 2022, 14(2): 229. |
| 109 | ÖHNSTEDT E, VÅGESJÖ E, FASTH A, et al. Engineered bacteria to accelerate wound healing: an adaptive, randomised, double-blind, placebo-controlled, first-in-human phase 1 trial[J]. EClinicalMedicine, 2023, 60: 102014. |
| 110 | ZHAO X X, LI S J, DING J N, et al. Combination of an engineered Lactococcus lactis expressing CXCL12 with light-emitting diode yellow light as a treatment for scalded skin in mice[J]. Microbial Biotechnology, 2021, 14(5): 2090-2100. |
| 111 | LI L Y, YANG C, MA B L, et al. Hydrogel-encapsulated engineered microbial consortium as a photoautotrophic “living material” for promoting skin wound healing[J]. ACS Applied Materials & Interfaces, 2023, 15(5): 6536-6547. |
| 112 | LU Y F, LI H S, WANG J, et al. Engineering bacteria-activated multifunctionalized hydrogel for promoting diabetic wound healing[J]. Advanced Functional Materials, 2021, 31(48): 2105749. |
| 113 | MING Z Z, HAN L, BAO M Y, et al. Living bacterial hydrogels for accelerated infected wound healing[J]. Advanced Science, 2021, 8(24): 2102545. |
| 114 | MIERAU I, LEIJ P, VAN SWAM I, et al. Industrial-scale production and purification of a heterologous protein in Lactococcus lactis using the nisin-controlled gene expression system NICE: the case of lysostaphin[J]. Microbial Cell Factories, 2005, 4: 15. |
| 115 | LUBKOWICZ D, HO C L, HWANG I Y, et al. Reprogramming probiotic Lactobacillus reuteri as a biosensor for Staphylococcus aureus derived AIP-I detection[J]. ACS Synthetic Biology, 2018, 7(5): 1229-1237. |
| [1] | 吴柯, 罗家豪, 李斐然. 机器学习驱动的基因组规模代谢模型构建与优化[J]. 合成生物学, 2025, 6(3): 566-584. |
| [2] | 田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546. |
| [3] | 章益蜻, 刘高雯. 合成生物学视角下的基因功能探索与酵母工程菌株文库构建[J]. 合成生物学, 2025, 6(3): 685-700. |
| [4] | 黄怡, 司同, 陆安静. 生物制造标准体系建设的现状、问题与建议[J]. 合成生物学, 2025, 6(3): 701-714. |
| [5] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [6] | 张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356. |
| [7] | 张璐鸥, 徐丽, 胡晓旭, 杨滢. 合成生物学助力化妆品走进生物制造新时代[J]. 合成生物学, 2025, 6(2): 479-491. |
| [8] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| [9] | 韦灵珍, 王佳, 孙新晓, 袁其朋, 申晓林. 黄酮类化合物生物合成及其在化妆品中应用的研究[J]. 合成生物学, 2025, 6(2): 373-390. |
| [10] | 肖森, 胡立涛, 石智诚, 王发银, 余思婷, 堵国成, 陈坚, 康振. 可控分子量透明质酸的生物合成研究进展[J]. 合成生物学, 2025, 6(2): 445-460. |
| [11] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [12] | 左一萌, 张姣姣, 连佳长. 酿酒酵母使能技术在化妆品原料合成中的应用[J]. 合成生物学, 2025, 6(2): 233-253. |
| [13] | 汤传根, 王璟, 张烁, 张昊宁, 康振. 功能肽合成和挖掘策略研究进展[J]. 合成生物学, 2025, 6(2): 461-478. |
| [14] | 张萍, 张维娇, 胥睿睿, 李江华, 陈坚, 康振. 防晒化合物类菌孢素氨基酸的生物合成[J]. 合成生物学, 2025, 6(2): 306-319. |
| [15] | 黄姝涵, 马赫, 罗云孜. 生物合成红景天苷的研究进展[J]. 合成生物学, 2025, 6(2): 391-407. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||