合成生物学 ›› 2025, Vol. 6 ›› Issue (3): 547-565.DOI: 10.12211/2096-8280.2025-016
统计物理与人工智能驱动的蛋白质结构生物信息学
夏辰亮1, 张泽成2, 管星悦3, 唐乾元2
- 1.三江学院数理部,江苏 南京 210012
2.香港浸会大学物理系,香港 999077
3.南京大学物理学院,江苏 南京 210093
-
收稿日期:2025-03-17修回日期:2025-04-15出版日期:2025-06-30发布日期:2025-06-27 -
通讯作者:唐乾元 -
作者简介:夏辰亮 (1990—),男,博士,讲师。研究方向为蛋白质动力学的统计物理研究。 E-mail:xiacl1030@qq.com张泽成 (1992—),男,博士研究生。研究方向为蛋白质序列、结构与动力学的统计、AI蛋白质结构预测和生物复杂性。E-mail:zhzece@outlook.com唐乾元 (1989—),男,博士,助理教授。研究方向为数据驱动的生物复杂系统理论框架构建,通过深度融合机器学习、统计物理与高性能计算方法,研究包括蛋白质分子和大脑等不同时空尺度的生物复杂系统,揭示其内在的普适性组织原理与动力学规律。 E-mail:tangqy@hkbu.edu.hk -
基金资助:国家自然科学基金(12305052);江苏省高等学校自然科学研究项目(22KJD14005);香港研究资助局杰出青年学者计划(22302723);香港浸会大学资助项目(RC-FNRA-IG/22-23/SCI/03)
Protein structural bioinformatics empowered by statistical physics and artificial intelligence
XIA Chenliang1, ZHANG Zecheng2, GUAN Xingyue3, TANG Qianyuan2
- 1.Department of Mathematics and Physics,Sanjiang University,Nanjing 210012,Jiangsu,China
2.Department of Physics,Hong Kong Baptist University,Hong Kong 999077,China
3.School of Physics,Nanjing University,Nanjing 210093,Jiangsu,China
-
Received:2025-03-17Revised:2025-04-15Online:2025-06-30Published:2025-06-27 -
Contact:TANG Qianyuan
摘要:
结构生物信息学聚焦于生物分子的三维结构及其功能,蛋白质的结构是其核心研究对象。深度学习引发的蛋白质结构预测革命,特别是AlphaFold2的突破,实现了仅凭氨基酸序列即可达到原子精度的蛋白质结构预测,从根本上重构了该领域的数据生态。统计物理学与大数据分析方法的深度融合,使研究者能够突破传统个案研究的局限,从海量数据中系统性揭示蛋白质设计的普适性规律。大规模蛋白质结构数据的积累为定量化研究蛋白质动力学中的长程关联及其与进化的对应关系奠定了重要基础,这不仅为理解蛋白质的结构、动力学、功能与进化提供了统一的理论框架,其揭示的普适规律与设计原则也为人工蛋白质设计提供了关键指导。在此基础上,基于AlphaFold数据库的跨物种蛋白质结构对比统计分析,突显了数据驱动方法在揭示蛋白质进化过程中随生物复杂性增加而呈现的普适统计规律方面的核心作用,为理解生命进化的分子机制提供了全新视角。鉴于蛋白质功能的实现往往依赖于多种构象状态间的动态转换,蛋白质动力学的精确预测已成为当前研究的核心方向。统计物理与人工智能相结合的研究范式将持续引领蛋白质科学的创新发展,通过提升高通量筛选和理性设计效率,加速从基础发现到实际应用的转化,为合成生物学、精准医学等领域开辟新的可能性。
中图分类号:
引用本文
夏辰亮, 张泽成, 管星悦, 唐乾元. 统计物理与人工智能驱动的蛋白质结构生物信息学[J]. 合成生物学, 2025, 6(3): 547-565.
XIA Chenliang, ZHANG Zecheng, GUAN Xingyue, TANG Qianyuan. Protein structural bioinformatics empowered by statistical physics and artificial intelligence[J]. Synthetic Biology Journal, 2025, 6(3): 547-565.
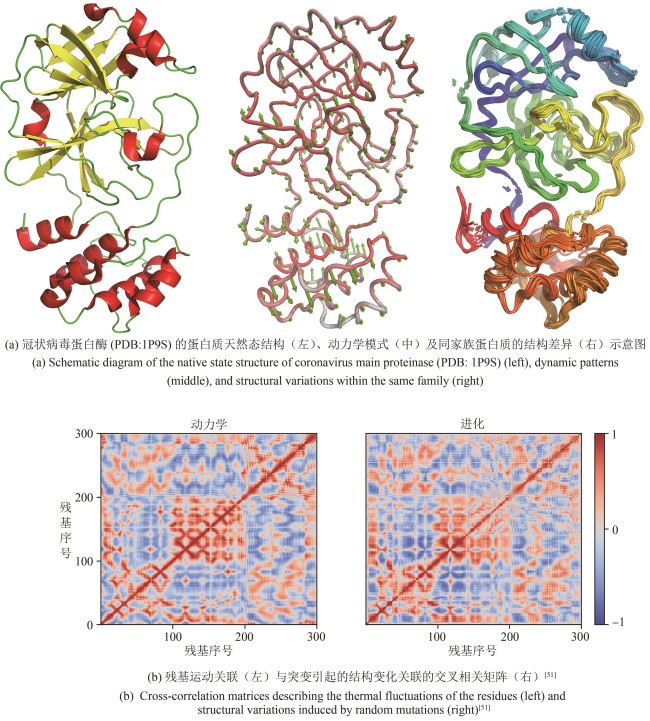
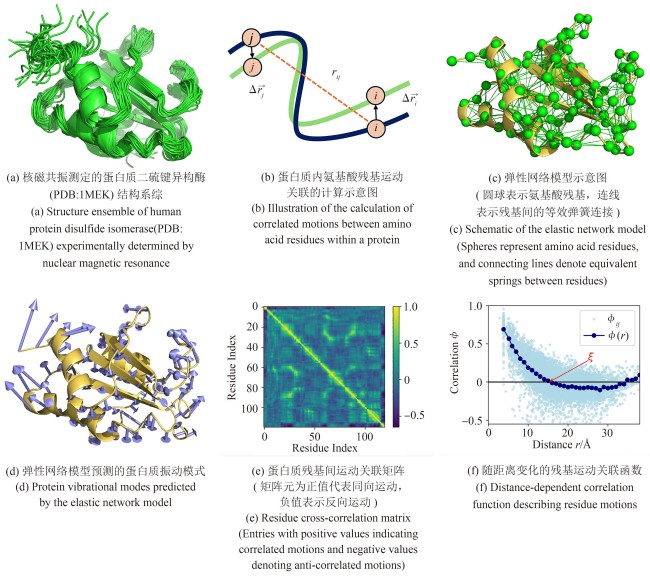
图2 关联分析方法在蛋白质结构动力学及突变引起的结构变化研究中的应用
Fig. 2 Application of correlation analysis in protein structural dynamics and mutation-induced structural variations
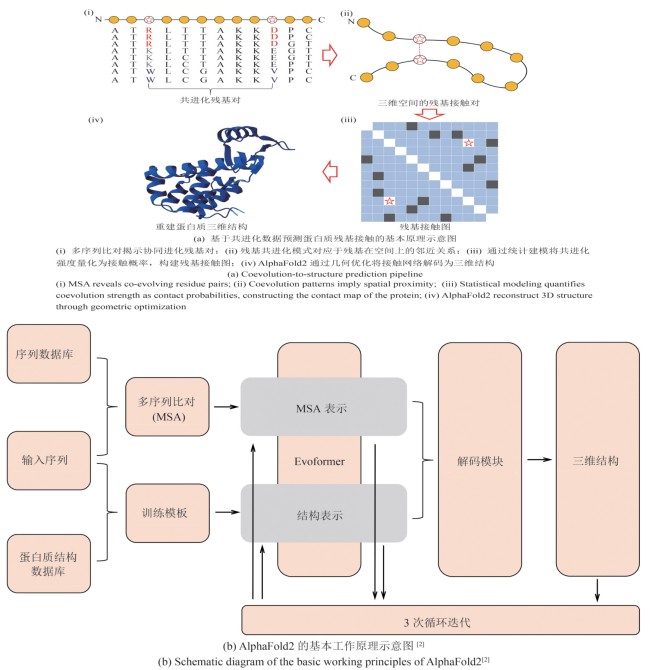
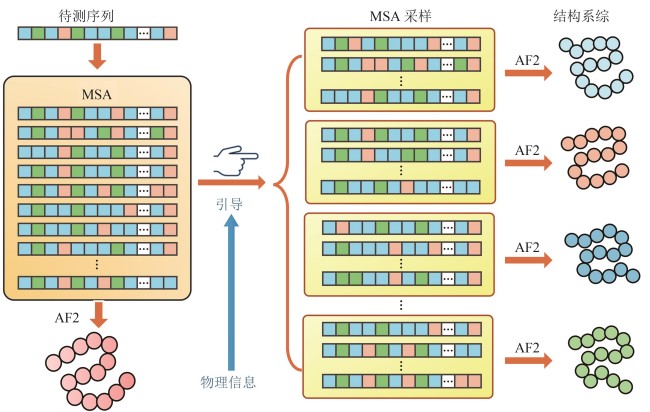
图3 基于共进化的残基接触预测与AlphaFold2蛋白质结构预测模型架构示意图
Fig. 3 Schematic illustration of the coevolution-based residue contact prediction and model architecture of AlphaFold2 for protein structure prediction
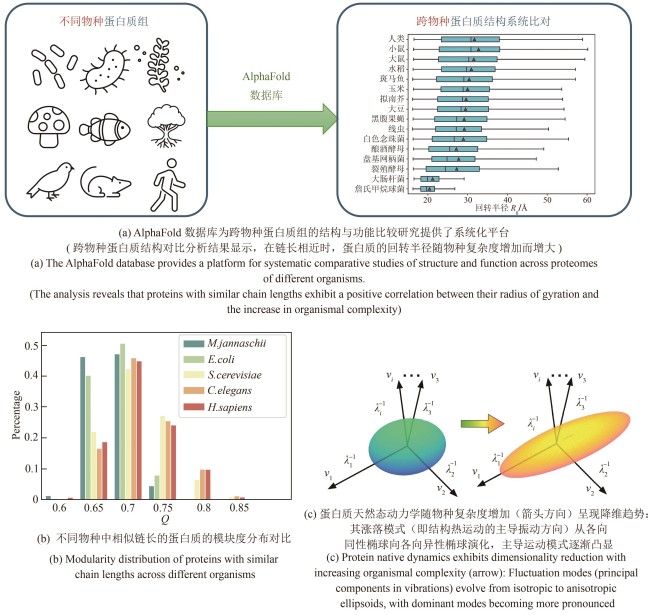
图4 基于AlphaFold数据库研究不同复杂度物种体内蛋白质结构与动力学的统计规律[100]
Fig. 4 Statistical trends in protein structure and dynamics across organisms of varying complexity: an analysis based on the AlphaFold database [100]
| 1 | CHOU K C. Structural bioinformatics and its impact to biomedical science[J]. Current Medicinal Chemistry, 2004, 11(16): 2105-2134. |
| 2 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 3 | VARADI M, ANYANGO S, DESHPANDE M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models[J]. Nucleic Acids Research, 2022, 50(D1): D439-D444. |
| 4 | KORTEMME T. De novo protein design-from new structures to programmable functions[J]. Cell, 2024, 187(3): 526-544. |
| 5 | HOPF T A, COLWELL L J, SHERIDAN R, et al. Three-dimensional structures of membrane proteins from genomic sequencing[J]. Cell, 2012, 149(7): 1607-1621. |
| 6 | MORA T, BIALEK W. Are biological systems poised at criticality?[J]. Journal of Statistical Physics, 2011, 144(2): 268-302. |
| 7 | HALILOGLU T, BAHAR I. Adaptability of protein structures to enable functional interactions and evolutionary implications[J]. Current Opinion in Structural Biology, 2015, 35: 17-23. |
| 8 | HALABI N, RIVOIRE O, LEIBLER S, et al. Protein sectors: evolutionary units of three-dimensional structure[J]. Cell, 2009, 138(4): 774-786. |
| 9 | NUSSINOV R, TSAI C J. Allostery in disease and in drug discovery[J]. Cell, 2013, 153(2): 293-305. |
| 10 | ORENGO C A, TODD A E, THORNTON J M. From protein structure to function[J]. Current Opinion in Structural Biology, 1999, 9(3): 374-382. |
| 11 | KARPLUS M, MCCAMMON J A. Molecular dynamics simulations of biomolecules[J]. Nature Structural Biology, 2002, 9(9): 646-652. |
| 12 | FRAUENFELDER H, CHEN G, BERENDZEN J, et al. A unified model of protein dynamics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(13): 5129-5134. |
| 13 | BOEHR D D, NUSSINOV R, WRIGHT P E. The role of dynamic conformational ensembles in biomolecular recognition[J]. Nature Chemical Biology, 2009, 5(11): 789-796. |
| 14 | HENZLER-WILDMAN K, KERN D. Dynamic personalities of proteins[J]. Nature, 2007, 450(7172): 964-972. |
| 15 | DE GENNES P G. Soft matter[J]. Science, 1992, 256(5056): 495-497. |
| 16 | CHANGEUX J P, CHRISTOPOULOS A. Allosteric modulation as a unifying mechanism for receptor function and regulation[J]. Cell, 2016, 166(5): 1084-1102. |
| 17 | KAY L E. NMR studies of protein structure and dynamics[J]. Journal of Magnetic Resonance, 2005, 173(2): 193-207. |
| 18 | XIE T, SALEH T, ROSSI P, et al. Conformational states dynamically populated by a kinase determine its function[J]. Science, 2020, 370(6513): eabc2754. |
| 19 | FRASER J S, VAN DEN BEDEM H, SAMELSON A J, et al. Accessing protein conformational ensembles using room-temperature X-ray crystallography[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(39): 16247-16252. |
| 20 | MERK A, BARTESAGHI A, BANERJEE S, et al. Breaking cryo-EM resolution barriers to facilitate drug discovery[J]. Cell, 2016, 165(7): 1698-1707. |
| 21 | FRANK J. Time-resolved cryo-electron microscopy: recent progress[J]. Journal of Structural Biology, 2017, 200(3): 303-306. |
| 22 | HARDER O F, BARRASS S V, DRABBELS M, et al. Fast viral dynamics revealed by microsecond time-resolved cryo-EM[J]. Nature Communications, 2023, 14: 5649. |
| 23 | KALTASHOV I A, BOBST C E, ABZALIMOV R R. Mass spectrometry-based methods to study protein architecture and dynamics[J]. Protein Science, 2013, 22(5): 530-544. |
| 24 | LENTO C, WILSON D J. Subsecond time-resolved mass spectrometry in dynamic structural biology[J]. Chemical Reviews, 2022, 122(8): 7624-7646. |
| 25 | BENKOVIC S J, HAMMES-SCHIFFER S. A perspective on enzyme catalysis[J]. Science, 2003, 301(5637): 1196-1202. |
| 26 | BAHAR I, LEZON T R, YANG L W, et al. Global dynamics of proteins: bridging between structure and function[J]. Annual Review of Biophysics, 2010, 39: 23-42. |
| 27 | BAHAR I, RADER A J. Coarse-grained normal mode analysis in structural biology[J]. Current Opinion in Structural Biology, 2005, 15(5): 586-592. |
| 28 | TIRION M M. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis[J]. Physical Review Letters, 1996, 77(9): 1905-1908. |
| 29 | TANG Q Y, KANEKO K. Long-range correlation in protein dynamics: confirmation by structural data and normal mode analysis[J]. PLoS Computational Biology, 2020, 16(2): e1007670. |
| 30 | REUVENI S, GRANEK R, KLAFTER J. Proteins: coexistence of stability and flexibility[J]. Physical Review Letters, 2008, 100(20): 208101. |
| 31 | TANG Q-Y, HATAKEYAMA T S, KANEKO K. Functional sensitivity and mutational robustness of proteins[J]. Physical Review Research, 2020, 2(3): 033452. |
| 32 | HU X H, HONG L, DEAN SMITH M, et al. The dynamics of single protein molecules is non-equilibrium and self-similar over thirteen decades in time[J]. Nature Physics, 2016, 12(2): 171-174. |
| 33 | TANG Q Y, ZHANG Y Y, WANG J, et al. Critical fluctuations in the native state of proteins[J]. Physical Review Letters, 2017, 118(8): 088102. |
| 34 | NEWMAN M E J. Modularity and community structure in networks[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(23): 8577-8582. |
| 35 | EISENMESSER E Z, MILLET O, LABEIKOVSKY W, et al. Intrinsic dynamics of an enzyme underlies catalysis[J]. Nature, 2005, 438(7064): 117-121. |
| 36 | STEIN A, FOWLER D M, HARTMANN-PETERSEN R, et al. Biophysical and mechanistic models for disease-causing protein variants[J]. Trends in Biochemical Sciences, 2019, 44(7): 575-588. |
| 37 | FRAUENFELDER H, SLIGAR S G, WOLYNES P G. The energy landscapes and motions of proteins[J]. Science, 1991, 254(5038): 1598-1603. |
| 38 | ZUCKERKANDL E, PAULING L. Evolutionary divergence and convergence in proteins[J]. Evolving Genes and Proteins, 1965: 97-166. |
| 39 | TAMA F, SANEJOUAND Y H. Conformational change of proteins arising from normal mode calculations[J]. Protein Engineering, 2001, 14(1): 1-6. |
| 40 | FACCO E, PAGNANI A, RUSSO E T, et al. The intrinsic dimension of protein sequence evolution[J]. PLoS Computational Biology, 2019, 15(4): e1006767. |
| 41 | LIU Y, BAHAR I. Sequence evolution correlates with structural dynamics[J]. Molecular Biology and Evolution, 2012, 29(9): 2253-2263. |
| 42 | TOKURIKI N, TAWFIK D S. Protein dynamism and evolvability[J]. Science, 2009, 324(5924): 203-207. |
| 43 | ILLERGÅRD K, ARDELL D H, ELOFSSON A. Structure is three to ten times more conserved than sequence: a study of structural response in protein cores[J]. Proteins: Structure, Function, and Bioinformatics, 2009, 77(3): 499-508. |
| 44 | WORTH C L, GONG S, BLUNDELL T L. Structural and functional constraints in the evolution of protein families[J]. Nature Reviews Molecular Cell Biology, 2009, 10(10): 709-720. |
| 45 | LIBERLES D A, TEICHMANN S A, BAHAR I, et al. The interface of protein structure, protein biophysics, and molecular evolution[J]. Protein Science, 2012, 21(6): 769-785. |
| 46 | ECHAVE J, WILKE C O. Biophysical models of protein evolution: understanding the patterns of evolutionary sequence divergence[J]. Annual Review of Biophysics, 2017, 46: 85-103. |
| 47 | BLOOM J D, LABTHAVIKUL S T, OTEY C R, et al. Protein stability promotes evolvability[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(15): 5869-5874. |
| 48 | SEROHIJOS A W R, SHAKHNOVICH E I. Merging molecular mechanism and evolution: theory and computation at the interface of biophysics and evolutionary population genetics[J]. Current Opinion in Structural Biology, 2014, 26: 84-91. |
| 49 | DRUMMOND D A, WILKE C O. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution[J]. Cell, 2008, 134(2): 341-352. |
| 50 | HOLM L. Dali server: structural unification of protein families[J]. Nucleic Acids Research, 2022, 50(W1): W210-W215. |
| 51 | TANG Q Y, KANEKO K. Dynamics-evolution correspondence in protein structures[J]. Physical Review Letters, 2021, 127(9): 098103. |
| 52 | ECHAVE J, SPIELMAN S J, WILKE C O. Causes of evolutionary rate variation among protein sites[J]. Nature Reviews Genetics, 2016, 17(2): 109-121. |
| 53 | MORCOS F, PAGNANI A, LUNT B, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(49): E1293-E1301. |
| 54 | HENZLER-WILDMAN K A, LEI M, THAI V, et al. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis[J]. Nature, 2007, 450(7171): 913-916. |
| 55 | GRANT B J, GORFE A A, MCCAMMON J A. Large conformational changes in proteins: signaling and other functions[J]. Current Opinion in Structural Biology, 2010, 20(2): 142-147. |
| 56 | NUSSINOV R, TSAI C J, LIU J. Principles of allosteric interactions in cell signaling[J]. Journal of the American Chemical Society, 2014, 136(51): 17692-17701. |
| 57 | MARSH J A, TEICHMANN S A. parallel dynamics and evolution: protein conformational fluctuations and assembly reflect evolutionary changes in sequence and structure[J]. BioEssays, 2014, 36(2): 209-218. |
| 58 | YANG L W, BAHAR I. Coupling between catalytic site and collective dynamics: a requirement for mechanochemical activity of enzymes[J]. Structure, 2005, 13(6): 893-904. |
| 59 | MAGUID S, FERNANDEZ-ALBERTI S, ECHAVE J. Evolutionary conservation of protein vibrational dynamics[J]. Gene, 2008, 422(1-2): 7-13. |
| 60 | TÓTH-PETRÓCZY Á, TAWFIK D S. The robustness and innovability of protein folds[J]. Current Opinion in Structural Biology, 2014, 26: 131-138. |
| 61 | LI H, TANG C, WINGREEN N S. Nature of driving force for protein folding: a result from analyzing the statistical potential[J]. Physical Review Letters, 1997, 79(4): 765-768. |
| 62 | ENGLAND J L, SHAKHNOVICH E I. Structural determinant of protein designability[J]. Physical Review Letters, 2003, 90(21): 218101. |
| 63 | BURLEY S K, BHATT R, BHIKADIYA C, et al. Updated resources for exploring experimentally-determined PDB structures and Computed Structure Models at the RCSB Protein Data Bank[J]. Nucleic Acids Research, 2025, 53(D1): D564-D574. |
| 64 | The UniProt Consortium. UniProt: the universal protein knowledgebase in 2023[J]. Nucleic Acids Research, 2023, 51(D1): D523-D531. |
| 65 | CHOTHIA C, LESK A M. The relation between the divergence of sequence and structure in proteins[J]. The EMBO Journal, 1986, 5(4): 823-826. |
| 66 | KRYSHTAFOVYCH A, SCHWEDE T, TOPF M, et al. Critical assessment of methods of protein structure prediction (CASP)-Round XIV[J]. Proteins: Structure, Function, and Bioinformatics, 2021, 89(12): 1607-1617. |
| 67 | FISER A, ŠALI A. Modeller: generation and refinement of homology-based protein structure models[J]. Methods in Enzymology, 2003, 374: 461-491. |
| 68 | WATERHOUSE A, BERTONI M, BIENERT S, et al. SWISS-MODEL: homology modelling of protein structures and complexes[J]. Nucleic Acids Research, 2018, 46(W1): W296-W303. |
| 69 | LAU K F, DILL K A. A lattice statistical mechanics model of the conformational and sequence spaces of proteins[J]. Macromolecules, 1989, 22(10): 3986-3997. |
| 70 | GO N. Theoretical studies of protein folding[J]. Annual Review of Biophysics and Bioengineering, 1983, 12: 183-210. |
| 71 | SIMONS K T, KOOPERBERG C, HUANG E, et al. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions[J]. Journal of Molecular Biology, 1997, 268(1): 209-225. |
| 72 | BRADLEY P, MISURA K M S, BAKER D. Toward high-resolution de novo structure prediction for small proteins[J]. Science, 2005, 309(5742): 1868-1871. |
| 73 | MIRDITA M, SCHÜTZE K, MORIWAKI Y, et al. ColabFold: making protein folding accessible to all[J]. Nature Methods, 2022, 19(6): 679-682. |
| 74 | AHDRITZ G, BOUATTA N, FLORISTEAN C, et al. OpenFold: retraining AlphaFold2 yields new insights into its learning mechanisms and capacity for generalization[J]. Nature Methods, 2024, 21(8): 1514-1524. |
| 75 | BAEK M, DIMAIO F, ANISHCHENKO I, et al. Accurate prediction of protein structures and interactions using a three-track neural network[J]. Science, 2021, 373(6557): 871-876. |
| 76 | BAEK M, BAKER D. Deep learning and protein structure modeling[J]. Nature Methods, 2022, 19(1): 13-14. |
| 77 | LIN Z M, AKIN H, RAO R, et al. Evolutionary-scale prediction of atomic-level protein structure with a language model[J]. Science, 2023, 379(6637): 1123-1130. |
| 78 | TUNYASUVUNAKOOL K, ADLER J, WU Z, et al. Highly accurate protein structure prediction for the human proteome[J]. Nature, 2021, 596(7873): 590-596. |
| 79 | WEIGT M, WHITE R A, SZURMANT H, et al. Identification of direct residue contacts in protein-protein interaction by message passing[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(1): 67-72. |
| 80 | SENIOR A W, EVANS R, JUMPER J, et al. Improved protein structure prediction using potentials from deep learning[J]. Nature, 2020, 577(7792): 706-710. |
| 81 | ABRAMSON J, ADLER J, DUNGER J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3[J]. Nature, 2024, 630(8016): 493-500. |
| 82 | WANG L, WEN Z H, LIU S W, et al. Overview of AlphaFold2 and breakthroughs in overcoming its limitations[J]. Computers in Biology and Medicine, 2024, 176: 108620. |
| 83 | YANG Z Y, ZENG X X, ZHAO Y, et al. AlphaFold2 and its applications in the fields of biology and medicine[J]. Signal Transduction and Targeted Therapy, 2023, 8: 115. |
| 84 | JUMPER J, EVANS R, PRITZEL A, et al. Applying and improving AlphaFold at CASP14[J]. Proteins: Structure, Function, and Bioinformatics, 2021, 89(12): 1711-1721. |
| 85 | XIA Y H, ZHAO K L, LIU D, et al. Multi-domain and complex protein structure prediction using inter-domain interactions from deep learning[J]. Communications Biology, 2023, 6: 1221. |
| 86 | SHOR B, SCHNEIDMAN-DUHOVNY D. CombFold: predicting structures of large protein assemblies using a combinatorial assembly algorithm and AlphaFold2[J]. Nature Methods, 2024, 21(3): 477-487. |
| 87 | TESEI G, TROLLE A I, JONSSON N, et al. Conformational ensembles of the human intrinsically disordered proteome[J]. Nature, 2024, 626(8000): 897-904. |
| 88 | EVANS R, O'NEILL M, PRITZEL A, et al. Protein complex prediction with AlphaFold-Multimer[EB/OL]. bioRxiv, 2021, 2021.10. 04.463034[2025-01-15]. . |
| 89 | BRYANT P, POZZATI G, ZHU W S, et al. Predicting the structure of large protein complexes using AlphaFold and Monte Carlo tree search[J]. Nature Communications, 2022, 13: 6028. |
| 90 | LIU S W, ZHU T, REN M L, et al. Predicting mutational effects on protein-protein binding via a side-chain diffusion probabilistic model[C/OL]//Advances in Neural Information Processing Systems, 2023, 36: 48994-49005 [2025-01-15]. . |
| 91 | TANG Q Y. The mechanics of protein sweet spots[J/OL]. Nature Physics, 2025. (2025-03-28)[2025-03-29]. . |
| 92 | WEINREB E, MCBRIDE J M, SIEK M, et al. Enzymes as viscoelastic catalytic machines[J/OL]. Nature Physics, 2025. (2025-03-28)[2025-03-29]. . |
| 93 | MA W J, ZHANG S G, LI Z, et al. Enhancing protein function prediction performance by utilizing AlphaFold-predicted protein structures[J]. Journal of Chemical Information and Modeling, 2022, 62(17): 4008-4017. |
| 94 | VAN KEMPEN M, KIM S S, TUMESCHEIT C, et al. Fast and accurate protein structure search with Foldseek[J]. Nature Biotechnology, 2024, 42(2): 243-246. |
| 95 | BARRIO-HERNANDEZ I, YEO J, JÄNES J, et al. Clustering predicted structures at the scale of the known protein universe[J]. Nature, 2023, 622(7983): 637-645. |
| 96 | KIM W S, MIRDITA M, LEVY KARIN E, et al. Rapid and sensitive protein complex alignment with Foldseek-Multimer[J]. Nature Methods, 2025, 22(3): 469-472. |
| 97 | DURAIRAJ J, WATERHOUSE A M, METS T, et al. Uncovering new families and folds in the natural protein universe[J]. Nature, 2023, 622(7983): 646-653. |
| 98 | ALDERSON T R, PRITIŠANAC I, KOLARIĆ Đ, et al. Systematic identification of conditionally folded intrinsically disordered regions by AlphaFold2[J]. Proceedings of the National Academy of Sciences of the United States of America, 2023, 120(44): e2304302120. |
| 99 | THORNTON J M, LASKOWSKI R A, BORKAKOTI N. AlphaFold heralds a data-driven revolution in biology and medicine[J]. Nature Medicine, 2021, 27(10): 1666-1669. |
| 100 | TANG Q Y, REN W T, WANG J, et al. The statistical trends of protein evolution: a lesson from AlphaFold database[J]. Molecular Biology and Evolution, 2022, 39(10): msac197. |
| 101 | SATO T U, KANEKO K. Evolutionary dimension reduction in phenotypic space[J]. Physical Review Research, 2020, 2(1): 013197. |
| 102 | SAKATA A, KANEKO K. Dimensional reduction in evolving spin-glass model: correlation of phenotypic responses to environmental and mutational changes[J]. Physical Review Letters, 2020, 124(21): 218101. |
| 103 | KANEKO K. Constructing universal phenomenology for biological cellular systems: an idiosyncratic review on evolutionary dimensional reduction[J]. Journal of Statistical Mechanics: Theory and Experiment, 2024, 2024(2): 024002. |
| 104 | KARPLUS M, KURIYAN J. Molecular dynamics and protein function[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(19): 6679-6685. |
| 105 | AMBROGGIO X I, KUHLMAN B. Design of protein conformational switches[J]. Current Opinion in Structural Biology, 2006, 16(4): 525-530. |
| 106 | MONTEIRO DA SILVA G, CUI J Y, DALGARNO D C, et al. High-throughput prediction of protein conformational distributions with subsampled AlphaFold2[J]. Nature Communications, 2024, 15: 2464. |
| 107 | STEIN R A, MCHAOURAB H S. SPEACH_AF: sampling protein ensembles and conformational heterogeneity with AlphaFold2[J]. PLoS Computational Biology, 2022, 18(8): e1010483. |
| 108 | DEL ALAMO D, SALA D, MCHAOURAB H S, et al. Sampling alternative conformational states of transporters and receptors with AlphaFold2[J]. eLife, 2022, 11: e75751. |
| 109 | WAYMENT-STEELE H K, OJOAWO A, OTTEN R, et al. Predicting multiple conformations via sequence clustering and AlphaFold2[J]. Nature, 2024, 625(7996): 832-839. |
| 110 | HEO L, FEIG M. Multi-state modeling of G-protein coupled receptors at experimental accuracy[J]. Proteins: Structure, Function, and Bioinformatics, 2022, 90(11): 1873-1885. |
| 111 | SALA D, ENGELBERGER F, MCHAOURAB H S, et al. Modeling conformational states of proteins with AlphaFold[J]. Current Opinion in Structural Biology, 2023, 81: 102645. |
| 112 | SALA D, HILDEBRAND P W, MEILER J. Biasing AlphaFold2 to predict GPCRs and kinases with user-defined functional or structural properties[J]. Frontiers in Molecular Biosciences, 2023, 10: 1121962. |
| 113 | WOLYNES P G, ONUCHIC J N, THIRUMALAI D. Navigating the folding routes[J]. Science, 1995, 267(5204): 1619-1620. |
| 114 | BRYNGELSON J D, ONUCHIC J N, SOCCI N D, et al. Funnels, pathways, and the energy landscape of protein folding: a synthesis[J]. Proteins: Structure, Function, and Bioinformatics, 1995, 21(3): 167-195. |
| 115 | FERREIRO D U, KOMIVES E A, WOLYNES P G. Frustration in biomolecules[J]. Quarterly Reviews of Biophysics, 2014, 47(4): 285-363. |
| 116 | PARRA R G, SCHAFER N P, RADUSKY L G, et al. Protein Frustratometer 2: a tool to localize energetic frustration in protein molecules, now with electrostatics[J]. Nucleic Acids Research, 2016, 44(W1): W356-W360. |
| 117 | FERREIRO D U, HEGLER J A, KOMIVES E A, et al. Localizing frustration in native proteins and protein assemblies[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(50): 19819-19824. |
| 118 | LI W F, WOLYNES P G, TAKADA S. Frustration, specific sequence dependence, and nonlinearity in large-amplitude fluctuations of allosteric proteins[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(9): 3504-3509. |
| 119 | CHEN M C, CHEN X, SCHAFER N P, et al. Surveying biomolecular frustration at atomic resolution[J]. Nature Communications, 2020, 11: 5944. |
| 120 | GIANNI S, FREIBERGER M I, JEMTH P, et al. Fuzziness and frustration in the energy landscape of protein folding, function, and assembly[J]. Accounts of Chemical Research, 2021, 54(5): 1251-1259. |
| 121 | GUAN X Y, TANG Q Y, REN W T, et al. Predicting protein conformational motions using energetic frustration analysis and AlphaFold2[J]. Proceedings of the National Academy of Sciences of the United States of America, 2024, 121(35): e2410662121. |
| 122 | XIE T Y, SONG Z L, HUANG J. Conditioned protein structure prediction[J]. PRX Life, 2024, 2(4): 043001. |
| 123 | CHAKRAVARTY D, SCHAFER J W, CHEN E A, et al. AlphaFold predictions of fold-switched conformations are driven by structure memorization[J]. Nature Communications, 2024, 15: 7296. |
| 124 | BRYANT P, NOÉ F. Structure prediction of alternative protein conformations[J]. Nature Communications, 2024, 15: 7328. |
| 125 | ZHANG J, LIU S R, CHEN M Y, et al. Unsupervisedly prompting AlphaFold2 for accurate few-shot protein structure prediction[J]. Journal of Chemical Theory and Computation, 2023, 19(22): 8460-8471. |
| 126 | LEE C Y, HUBRICH D, VARGA J K, et al. Systematic discovery of protein interaction interfaces using AlphaFold and experimental validation[J]. Molecular Systems Biology, 2024, 20(2): 75-97. |
| 127 | GUO Z Y, LIU J, WANG Y L, et al. Diffusion models in bioinformatics and computational biology[J]. Nature Reviews Bioengineering, 2024, 2(2): 136-154. |
| 128 | WU K E, YANG K K, VAN DEN BERG R, et al. Protein structure generation via folding diffusion[J]. Nature Communications, 2024, 15: 1059. |
| 129 | PILLAI A, IDRIS A, PHILOMIN A, et al. De novo design of allosterically switchable protein assemblies[J]. Nature, 2024, 632(8026): 911-920. |
| 130 | ECKMANN J P, ROUGEMONT J, TLUSTY T. Colloquium: proteins: the physics of amorphous evolving matter[J]. Reviews of Modern Physics, 2019, 91(3): 031001. |
| 131 | CHENG J, NOVATI G, PAN J, et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense[J]. Science, 2023, 381(6664): eadg7492. |
| 132 | MARCHETTI F, MORONI E, PANDINI A, et al. Machine learning prediction of allosteric drug activity from molecular dynamics[J]. The Journal of Physical Chemistry Letters, 2021, 12(15): 3724-3732. |
| 133 | BAI Q F, LIU S, TIAN Y N, et al. Application advances of deep learning methods for de novo drug design and molecular dynamics simulation[J]. Wiley Interdisciplinary Reviews: Computational Molecular Science, 2022, 12(3): e1581. |
| 134 | PERRAKIS A, SIXMA T K. AI revolutions in biology: the joys and perils of AlphaFold[J]. EMBO Reports, 2021, 22(11): e54046. |
| 135 | MONZON V, HAFT D H, BATEMAN A. Folding the unfoldable: using AlphaFold to explore spurious proteins[J]. Bioinformatics Advances, 2022, 2(1): vbab043. |
| 136 | ANISHCHENKO I, PELLOCK S J, CHIDYAUSIKU T M, et al. De novo protein design by deep network hallucination[J]. Nature, 2021, 600(7889): 547-552. |
| 137 | YANG K K, WU Z, ARNOLD F H. Machine-learning-guided directed evolution for protein engineering[J]. Nature Methods, 2019, 16(8): 687-694. |
| 138 | BAYLY-JONES C, WHISSTOCK J C. Mining folded proteomes in the era of accurate structure prediction[J]. PLoS Computational Biology, 2022, 18(3): e1009930. |
| 139 | LIU X Y, XING J Y, FU H H, et al. Analyzing molecular dynamics trajectories thermodynamically through artificial intelligence[J]. Journal of Chemical Theory and Computation, 2024, 20(2): 665-676. |
| 140 | WANG T, HE X H, LI M Y, et al. Ab initio characterization of protein molecular dynamics with AI2BMD[J]. Nature, 2024, 635(8040): 1019-1027. |
| 141 | BOLON D N, GRANT R A, BAKER T A, et al. Specificity versus stability in computational protein design[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(36): 12724-12729. |
| 142 | PECCATI F, ALUNNO-RUFINI S, JIMÉNEZ-OSÉS G. Accurate prediction of enzyme thermostabilization with Rosetta using AlphaFold ensembles[J]. Journal of Chemical Information and Modeling, 2023, 63(3): 898-909. |
| 143 | ZHU J, AVAKYAN N, KAKKIS A, et al. Protein assembly by design[J]. Chemical Reviews, 2021, 121(22): 13701-13796. |
| 144 | HAYES T, RAO R, AKIN H, et al. Simulating 500 million years of evolution with a language model[J]. Science, 2025, 387(6736): 850-858. |
| 145 | ROMERO P A, ARNOLD F H. Exploring protein fitness landscapes by directed evolution[J]. Nature Reviews Molecular Cell Biology, 2009, 10(12): 866-876. |
| 146 | JIANG K Y, YAN Z Q, DI BERNARDO M, et al. Rapid in silico directed evolution by a protein language model with EVOLVEpro[J]. Science, 2025, 387(6732): eadr6006. |
| 147 | KING R D, ROWLAND J, OLIVER S G, et al. The automation of science[J]. Science, 2009, 324(5923): 85-89. |
| 148 | SAVINOV A, SWANSON S, KEATING A E, et al. High-throughput discovery of inhibitory protein fragments with AlphaFold[J]. Biophysical Journal, 2024, 123(3): 55A-56A. |
| [1] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [2] | 雷茹, 陶慧, 刘天罡. 基因组深度挖掘驱动微生物萜类化合物高效发现[J]. 合成生物学, 2024, 5(3): 507-526. |
| [3] | 奚萌宇, 胡逸灵, 顾玉诚, 戈惠明. 基因组挖掘指导天然药物分子的发现[J]. 合成生物学, 2024, 5(3): 447-473. |
| [4] | 朱景勇, 李钧翔, 李旭辉, 张瑾, 毋文静. 深度学习在基于序列的蛋白质互作预测中的应用进展[J]. 合成生物学, 2024, 5(1): 88-106. |
| [5] | 王晟, 王泽琛, 陈威华, 陈珂, 彭向达, 欧发芬, 郑良振, 孙瑨原, 沈涛, 赵国屏. 基于人工智能和计算生物学的合成生物学元件设计[J]. 合成生物学, 2023, 4(3): 422-443. |
| [6] | 康里奇, 谈攀, 洪亮. 人工智能时代下的酶工程[J]. 合成生物学, 2023, 4(3): 524-534. |
| [7] | 孟巧珍, 郭菲. “可折叠性”在酶智能设计改造中的应用研究——以AlphaFold2为例[J]. 合成生物学, 2023, 4(3): 571-589. |
| [8] | 陈志航, 季梦麟, 戚逸飞. 人工智能蛋白质结构设计算法研究进展[J]. 合成生物学, 2023, 4(3): 464-487. |
| [9] | 赖奇龙, 姚帅, 查毓国, 白虹, 宁康. 微生物组生物合成基因簇发掘方法及应用前景[J]. 合成生物学, 2023, 4(3): 611-627. |
| [10] | 卞佳豪, 杨广宇. 人工智能辅助的蛋白质工程[J]. 合成生物学, 2022, 3(3): 429-444. |
| [11] | 于慧敏, 郑煜堃, 杜岩, 王苗苗, 梁有向. 合成生物学研究中的微生物启动子工程策略[J]. 合成生物学, 2021, 2(4): 598-611. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||