合成生物学 ›› 2025, Vol. 6 ›› Issue (3): 532-546.DOI: 10.12211/2096-8280.2024-083
合成基因回路面临的细胞“经济学窘境”
田晓军, 张日新
- 亚利桑那州立大学生物与健康系统工程学院,美国 亚利桑那州 坦佩 85281
-
收稿日期:2024-11-27修回日期:2025-02-19出版日期:2025-06-30发布日期:2025-06-27 -
通讯作者:田晓军 -
作者简介:田晓军 (1984—),男,博士生导师。研究方向为定量生物学、系统生物学、合成生物学,目前研究是优化人工合成基因回路设计及其应用。E-mail:Xiaojun.Tian@asu.edu
“Economics Paradox” with cells in synthetic gene circuits
TIAN Xiao-jun, ZHANG Rixin
- School of Biological and Health Systems Engineering,Arizona State University,Tempe 85281,Arizona,USA
-
Received:2024-11-27Revised:2025-02-19Online:2025-06-30Published:2025-06-27 -
Contact:TIAN Xiao-jun
摘要:
在合成生物学中,基因模块是执行生物功能的核心元件。“模块性”是指已知基因元件在被拼装为目的基因回路后仍能保持其功能相对独立的特性。不同于传统工程学独立且稳定的特性,基因回路存在于动态变化的细胞环境中,其功能蛋白的表达效率高度依赖于胞内资源。有限的资源分配使得基因回路面临胞内资源的约束挑战,导致基因回路的模块性丧失。恢复基因回路的模块性有助于构建普适的生命系统理论模型,推动人工生命体系的智能化发展。近年来,有关资源竞争如何重塑基因回路表现的研究逐渐增多,这些研究加深了对潜在作用机制的理解,并推动了基因回路设计的优化。本综述系统阐述了细胞资源竞争现象对基因回路功能的影响,包括基因回路噪声的改变,基因模块的耦合关系,以及赢者通吃的涌现性。同时,对现有控制策略进行了全面归纳,包括细胞资源的正交化设计,单基因模块的资源调控以及多基因模块的统筹化控制。随着合成生物学的快速发展,人工设计的基因回路在结构和功能上会变得更加复杂。这一趋势预示着未来的研究重点将不再局限于简单的资源竞争控制体系,而需要向更大规模的研究范畴拓展。与此同时,研究方向应从基础研究探索延伸至实际应用,最终实现精确可控的人工生命体系的构建。
中图分类号:
引用本文
田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546.
TIAN Xiao-jun, ZHANG Rixin. “Economics Paradox” with cells in synthetic gene circuits[J]. Synthetic Biology Journal, 2025, 6(3): 532-546.
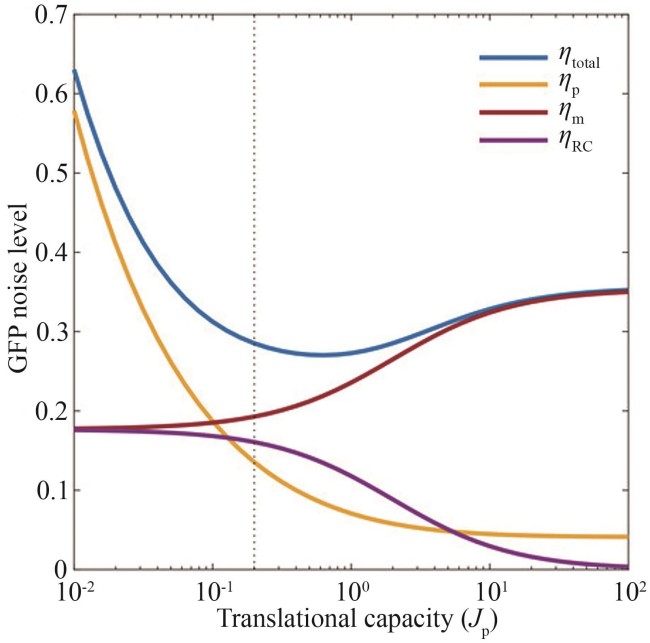
图2 总体蛋白噪声的解析解曲线[34](ηtotal—蛋白总噪声;ηp—蛋白生成或降解噪声;ηm—mRNA波动噪声;ηRC—资源竞争噪声)
Fig. 2 Analytical solutions of the total protein noise[34](ηtotal—total protein noise; ηp—birth/death of protein noise; ηm—mRNA fluctuating noise; ηRC—resource competition noise)
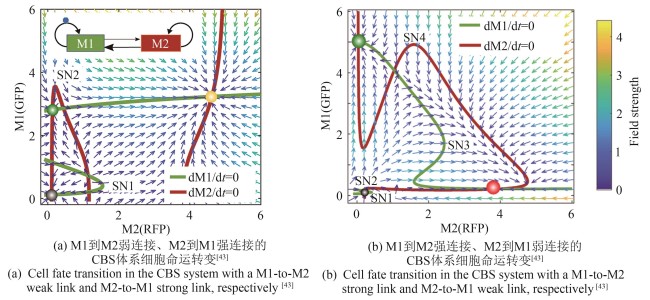
图4 基于两模块的不同连接强度,级联双稳态开关(CBS)呈现出不同的两种细胞命运转变途径[43]
Fig. 4 Cascaded bistable switch circuit demonstrates two different paths for cell fate transition associated with the strength of links between the two modules[43]
| 1 | MOE-BEHRENS G H G, DAVIS R, HAYNES K A. Preparing synthetic biology for the world[J]. Frontiers in Microbiology, 2013, 4: 5. |
| 2 | WURTZEL E T, VICKERS C E, HANSON A D, et al. Revolutionizing agriculture with synthetic biology[J]. Nature Plants, 2019, 5(12): 1207-1210. |
| 3 | MENG F K, ELLIS T. The second decade of synthetic biology: 2010—2020[J]. Nature Communications, 2020, 11(1): 5174. |
| 4 | NGUYEN P Q, HUANG X N, COLLINS D S, et al. Harnessing synthetic biology to enhance ocean health[J]. Trends in Biotechnology, 2023, 41(7): 860-874. |
| 5 | TANG T C, AN B L, HUANG Y Y, et al. Materials design by synthetic biology[J]. Nature Reviews Materials, 2021, 6(4): 332-350. |
| 6 | BORKOWSKI O, CERONI F, STAN G B, et al. Overloaded and stressed: whole-cell considerations for bacterial synthetic biology[J]. Current Opinion in Microbiology, 2016, 33: 123-130. |
| 7 | SHAKIBA N, JONES R D, WEISS R, et al. Context-aware synthetic biology by controller design: engineering the mammalian cell[J]. Cell Systems, 2021, 12(6): 561-592. |
| 8 | LIAO C, BLANCHARD A E, LU T. An integrative circuit-host modelling framework for predicting synthetic gene network behaviours[J]. Nature Microbiology, 2017, 2(12): 1658-1666. |
| 9 | DEL VECCHIO D. Modularity, context-dependence, and insulation in engineered biological circuits[J]. Trends in Biotechnology, 2015, 33(2): 111-119. |
| 10 | BOO A, ELLIS T, STAN G B. Host-aware synthetic biology[J]. Current Opinion in Systems Biology, 2019, 14: 66-72. |
| 11 | ILIA K, DEL VECCHIO D. Squaring a circle: to what extent are traditional circuit analogies impeding synthetic biology?[J]. GEN Biotechnology, 2022, 1(2): 150-155. |
| 12 | ŞIMŞEK E, YAO Y, LEE D, et al. Toward predictive engineering of gene circuits[J]. Trends in Biotechnology, 2023, 41(6): 760-768. |
| 13 | BREMER H, DENNIS P P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates[J]. EcoSal Plus, 2008, 3(1): 10.1128/ecosal.5.2.3. |
| 14 | VIND J, SØRENSEN M A, RASMUSSEN M D, et al. Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes. Expression from reporter genes does not always reflect functional mRNA levels[J]. Journal of Molecular Biology, 1993, 231(3): 678-688. |
| 15 | CHURCHWARD G, BREMER H, YOUNG R. Transcription in bacteria at different DNA concentrations[J]. Journal of Bacteriology, 1982, 150(2): 572-581. |
| 16 | STOEBEL D M, DEAN A M, DYKHUIZEN D E. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products[J]. Genetics, 2008, 178(3): 1653-1660. |
| 17 | CARBONELL-BALLESTERO M, GARCIA-RAMALLO E, MONTAÑEZ R, et al. Dealing with the genetic load in bacterial synthetic biology circuits: convergences with the Ohm's law[J]. Nucleic Acids Research, 2016, 44(1): 496-507. |
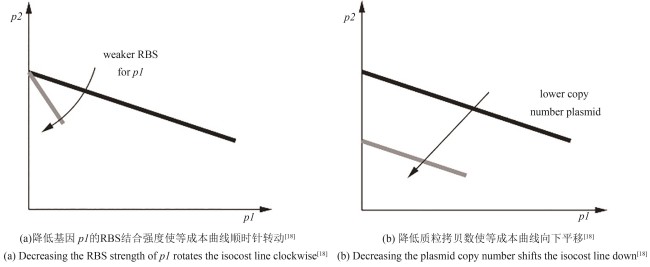
| 18 | GYORGY A, JIMÉNEZ J I, YAZBEK J, et al. Isocost lines describe the cellular economy of genetic circuits[J]. Biophysical Journal, 2015, 109(3): 639-646. |
| 19 | DEL VECCHIO D, QIAN Y L, MURRAY R M, et al. Future systems and control research in synthetic biology[J]. Annual Reviews in Control, 2018, 45: 5-17. |
| 20 | BASHOR C J, COLLINS J J. Insulating gene circuits from context by RNA processing[J]. Nature Biotechnology, 2012, 30(11): 1061-1062. |
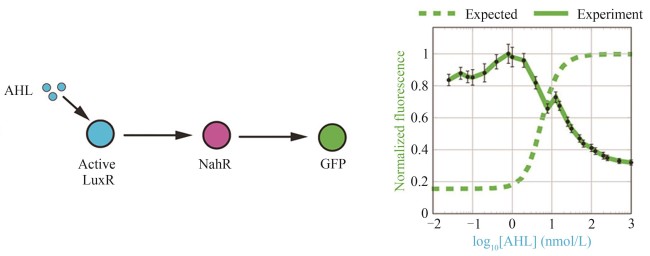
| 21 | QIAN Y L, HUANG H H, JIMÉNEZ J I, et al. Resource competition shapes the response of genetic circuits[J]. ACS Synthetic Biology, 2017, 6(7): 1263-1272. |
| 22 | CERONI F, ALGAR R, STAN G B, et al. Quantifying cellular capacity identifies gene expression designs with reduced burden[J]. Nature Methods, 2015, 12(5): 415-418. |
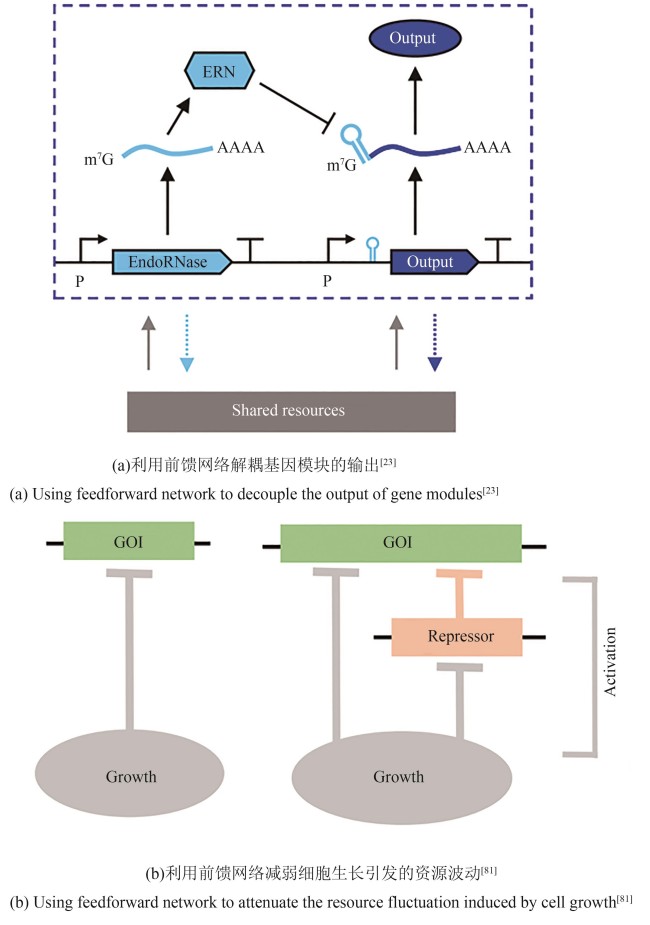
| 23 | JONES R D, QIAN Y L, SICILIANO V, et al. An endoribonuclease-based feedforward controller for decoupling resource-limited genetic modules in mammalian cells[J]. Nature Communications, 2020, 11(1): 5690. |
| 24 | DI BLASI R, PISANI M, TEDESCHI F, et al. Resource-aware construct design in mammalian cells[J]. Nature Communications, 2023, 14(1): 3576. |
| 25 | MORIYA T, YAMAOKA T, WAKAYAMA Y, et al. Comparison between effects of retroactivity and resource competition upon change in downstream reporter genes of synthetic genetic circuits[J]. Life, 2019, 9(1): 30. |
| 26 | CAMERON D E, COLLINS J J. Tunable protein degradation in bacteria[J]. Nature Biotechnology, 2014, 32(12): 1276-1281. |
| 27 | HERMSEN R, TANS S, WOLDE P R TEN. Transcriptional regulation by competing transcription factor modules[J]. PLoS Computational Biology, 2006, 2(12): e164. |
| 28 | DONG H J, NILSSON L, KURLAND C G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates[J]. Journal of Molecular Biology, 1996, 260(5): 649-663. |
| 29 | LOVE A M, NAIR N U. Specific codons control cellular resources and fitness[J]. Science Advances, 2024, 10(8): eadk3485. |
| 30 | COOKSON N A, MATHER W H, DANINO T, et al. Queueing up for enzymatic processing: correlated signaling through coupled degradation[J]. Molecular Systems Biology, 2011, 7: 561. |
| 31 | BUTZIN N C, HOCHENDONER P, OGLE C T, et al. Entrainment of a bacterial synthetic gene oscillator through proteolytic queueing[J]. ACS Synthetic Biology, 2017, 6(3): 455-462. |
| 32 | PAULSSON J. Models of stochastic gene expression[J]. Physics of Life Reviews, 2005, 2(2): 157-175. |
| 33 | PAULSSON J. Summing up the noise in gene networks[J]. Nature, 2004, 427(6973): 415-418. |
| 34 | GOETZ H, STONE A, ZHANG R, et al. Double-edged role of resource competition in gene expression noise and control[J]. Advanced Genetics, 2022, 3(1): 2100050. |
| 35 | ALON U. Network motifs: theory and experimental approaches[J]. Nature Reviews Genetics, 2007, 8(6): 450-461. |
| 36 | SHEN-ORR S S, MILO R, MANGAN S, et al. Network motifs in the transcriptional regulation network of Escherichia coli [J]. Nature Genetics, 2002, 31(1): 64-68. |
| 37 | MILO R, SHEN-ORR S, ITZKOVITZ S, et al. Network motifs: simple building blocks of complex networks[J]. Science, 2002, 298(5594): 824-827. |
| 38 | WANG L, WALKER B L, IANNACCONE S, et al. Bistable switches control memory and plasticity in cellular differentiation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(16): 6638-6643. |
| 39 | VEENING J W, SMITS W K, KUIPERS O P. Bistability, epigenetics, and bet-hedging in bacteria[J]. Annual Review of Microbiology, 2008, 62: 193-210. |
| 40 | CHAKRABORTY P, GHOSH S. Emergent correlations in gene expression dynamics as footprints of resource competition[J]. The European Physical Journal E, 2021, 44(10): 131. |
| 41 | PARTCH C L, GREEN C B, TAKAHASHI J S. Molecular architecture of the mammalian circadian clock[J]. Trends in Cell Biology, 2014, 24(2): 90-99. |
| 42 | ITO H, MUTSUDA M, MURAYAMA Y, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus [J]. Proceedingsof the National Academy of Sciences of the United States of America, 2009, 106(33): 14168-14173. |
| 43 | ZHANG R, GOETZ H, MELENDEZ-ALVAREZ J, et al. Winner-takes-all resource competition redirects cascading cell fate transitions[J]. Nature Communications, 2021, 12(1): 853. |
| 44 | LIANG S T, XU Y C, DENNIS P, et al. mRNA composition and control of bacterial gene expression[J]. Journal of Bacteriology, 2000, 182(11): 3037-3044. |
| 45 | LIANG S T, BIPATNATH M, XU Y C, et al. Activities of constitutive promoters in Escherichia coli [J]. Journal of Molecular Biology, 1999, 292(1): 19-37. |
| 46 | KLUMPP S, HWA T. Growth-rate-dependent partitioning of RNA polymerases in bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(51): 20245-20250. |
| 47 | KLUMPP S, ZHANG Z G, HWA T. Growth rate-dependent global effects on gene expression in bacteria[J]. Cell, 2009, 139(7): 1366-1375. |
| 48 | ALEXANDER W A, MOSS B, FUERST T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor[J]. Journal of Virology, 1992, 66(5): 2934-2942. |
| 49 | CHAMBERLIN M, MCGRATH J, WASKELL L. New RNA polymerase from Escherichia coli infected with bacteriophage T7[J]. Nature, 1970, 228(5268): 227-231. |
| 50 | STUDIER F W, MOFFATT B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes[J]. Journal of Molecular Biology, 1986, 189(1): 113-130. |
| 51 | KUSHWAHA M, SALIS H M. A portable expression resource for engineering cross-species genetic circuits and pathways[J]. Nature Communications, 2015, 6: 7832. |
| 52 | EL-SAMAD H, KURATA H, DOYLE J C, et al. Surviving heat shock: control strategies for robustness and performance[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(8): 2736-2741. |
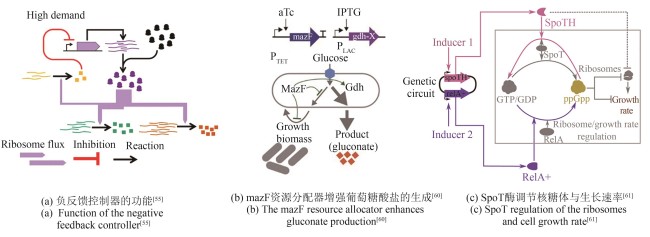
| 53 | SEGALL-SHAPIRO T H, MEYER A J, ELLINGTON A D, et al. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase[J]. Molecular Systems Biology, 2014, 10(7): 742. |
| 54 | DARLINGTON A P S, BATES D G. Architectures for combined transcriptional and translational resource allocation controllers[J]. Cell Systems, 2020, 11(4): 382-392. e9. |
| 55 | DARLINGTON A P S, KIM J, JIMÉNEZ J I, et al. Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes[J]. Nature Communications, 2018, 9(1): 695. |
| 56 | ORELLE C, CARLSON E D, SZAL T, et al. Protein synthesis by ribosomes with tethered subunits[J]. Nature, 2015, 524(7563): 119-124. |
| 57 | ALEKSASHIN N A, SZAL T, D’AQUINO A E, et al. A fully orthogonal system for protein synthesis in bacterial cells[J]. Nature Communications, 2020, 11(1): 1858. |
| 58 | DE JONG H, GEISELMANN J, ROPERS D. Resource reallocation in bacteria by reengineering the gene expression machinery[J]. Trends in Microbiology, 2017, 25(6): 480-493. |
| 59 | DARLINGTON A P S, KIM J, JIMÉNEZ J I, et al. Engineering translational resource allocation controllers: mechanistic models, design guidelines, and potential biological implementations[J]. ACS Synthetic Biology, 2018, 7(11): 2485-2496. |
| 60 | VENTURELLI O S, TEI M, BAUER S, et al. Programming mRNA decay to modulate synthetic circuit resource allocation[J]. Nature Communications, 2017, 8: 15128. |
| 61 | BARAJAS C, HUANG H H, GIBSON J, et al. Feedforward growth rate control mitigates gene activation burden[J]. Nature Communications, 2022, 13(1): 7054. |
| 62 | ZHU M L, DAI X F. Growth suppression by altered (p)ppGpp levels results from non-optimal resource allocation in Escherichia coli [J]. Nucleic Acids Research, 2019, 47(9): 4684-4693. |
| 63 | BÜKE F, GRILLI J, COSENTINO LAGOMARSINO M, et al. ppGpp is a bacterial cell size regulator[J]. Current Biology, 2022, 32(4): 870-877. e5. |
| 64 | MU H Y, HAN F, WANG Q, et al. Recent functional insights into the magic role of (p)ppGpp in growth control[J]. Computational and Structural Biotechnology Journal, 2023, 21: 168-175. |
| 65 | ROELL G W, ZHA J, CARR R R, et al. Engineering microbial consortia by division of labor[J]. Microbial Cell Factories, 2019, 18(1): 35. |
| 66 | TSOI R, WU F L, ZHANG C, et al. Metabolic division of labor in microbial systems[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2526-2531. |
| 67 | OVÁDI J, SAKS V. On the origin of intracellular compartmentation and organized metabolic systems[J]. Molecular and Cellular Biochemistry, 2004, 256-257(1-2): 5-12. |
| 68 | MAMPEL J, BUESCHER J M, MEURER G, et al. Coping with complexity in metabolic engineering[J]. Trends in Biotechnology, 2013, 31(1): 52-60. |
| 69 | CHOWDHURY C, SINHA S, CHUN S, et al. Diverse bacterial microcompartment organelles[J]. Microbiology and Molecular Biology Reviews, 2014, 78(3): 438-468. |
| 70 | YEATES T O, JORDA J, BOBIK T A. The shells of BMC-type microcompartment organelles in bacteria[J]. Journal of Molecular Microbiology and Biotechnology, 2013, 23(4-5): 290-299. |
| 71 | KERFELD C A, HEINHORST S, CANNON G C. Bacterial microcompartments[J]. Annual Review of Microbiology, 2010, 64: 391-408. |
| 72 | THOMMES M, WANG T Y, ZHAO Q, et al. Designing metabolic division of labor in microbial communities[J]. mSystems, 2019, 4(2): e00263-18. |
| 73 | LINDEMANN S R. A piece of the pie: engineering microbiomes by exploiting division of labor in complex polysaccharide consumption[J]. Current Opinion in Chemical Engineering, 2020, 30: 96-102. |
| 74 | ALNAHHAS R N, WINKLE J J, HIRNING A J, et al. Spatiotemporal dynamics of synthetic microbial consortia in microfluidic devices[J]. ACS Synthetic Biology, 2019, 8(9): 2051-2058. |
| 75 | KIM H J, BOEDICKER J Q, CHOI J W, et al. Defined spatial structure stabilizes a synthetic multispecies bacterial community[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(47): 18188-18193. |
| 76 | XU P. Dynamics of microbial competition, commensalism, and cooperation and its implications for coculture and microbiome engineering[J]. Biotechnology and Bioengineering, 2021, 118(1): 199-209. |
| 77 | SHOPERA T, HE L, OYETUNDE T, et al. Decoupling resource-coupled gene expression in living cells[J]. ACS Synthetic Biology, 2017, 6(8): 1596-1604. |
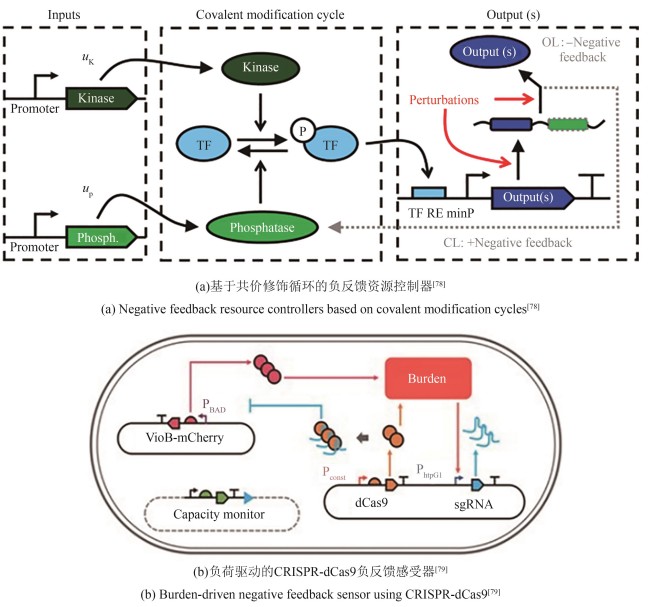
| 78 | JONES R D, QIAN Y L, ILIA K, et al. Robust and tunable signal processing in mammalian cells via engineered covalent modification cycles[J]. Nature Communications, 2022, 13(1): 1720. |
| 79 | CERONI F, BOO A, FURINI S, et al. Burden-driven feedback control of gene expression[J]. Nature Methods, 2018, 15(5): 387-393. |
| 80 | FREI T, CELLA F, TEDESCHI F, et al. Characterization and mitigation of gene expression burden in mammalian cells[J]. Nature Communications, 2020, 11(1): 4641. |
| 81 | STONE A, RIJAL S, ZHANG R, et al. Enhancing circuit stability under growth feedback with supplementary repressive regulation[J]. Nucleic Acids Research, 2024, 52(3): 1512-1521. |
| 82 | HUANG H H, QIAN Y L, DEL VECCHIO D. A quasi-integral controller for adaptation of genetic modules to variable ribosome demand[J]. Nature Communications, 2018, 9(1): 5415. |
| 83 | STONE A, RYAN J, TANG X, et al. Negatively competitive incoherent feedforward loops mitigate winner-take-all resource competition[J]. ACS Synthetic Biology, 2022, 11(12): 3986-3995. |
| 84 | STONE A, ZHANG R, TIAN X J. Coupling shared and tunable negative competition against winner-take-all resource competition via CRISPRi moieties[C/OL]//2021 American Control Conference (ACC). IEEE, 2021: 1-6. (2021-07-28)[2024-11-01]. . |
| 85 | CHAKRAVARTY S, ZHANG R, TIAN X J. Noise reduction in resource-coupled multi-module gene circuits through antithetic feedback control[J]. bioRxiv, 2024: 2024. 05. 24. 595570. |
| 86 | CHAKRAVARTY S, GUTTAL R, ZHANG R, et al. Mitigating winner-take-all resource competition through antithetic control mechanism[J]. ACS Synthetic Biology, 2024, 13(12): 4050-4060. |
| 87 | RAI K, WANG Y D, O’CONNELL R W, et al. Using machine learning to enhance and accelerate synthetic biology[J]. Current Opinion in Biomedical Engineering, 2024, 31: 100553. |
| [1] | 吴柯, 罗家豪, 李斐然. 机器学习驱动的基因组规模代谢模型构建与优化[J]. 合成生物学, 2025, 6(3): 566-584. |
| [2] | 章益蜻, 刘高雯. 合成生物学视角下的基因功能探索与酵母工程菌株文库构建[J]. 合成生物学, 2025, 6(3): 685-700. |
| [3] | 黄怡, 司同, 陆安静. 生物制造标准体系建设的现状、问题与建议[J]. 合成生物学, 2025, 6(3): 701-714. |
| [4] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [5] | 张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356. |
| [6] | 张璐鸥, 徐丽, 胡晓旭, 杨滢. 合成生物学助力化妆品走进生物制造新时代[J]. 合成生物学, 2025, 6(2): 479-491. |
| [7] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| [8] | 韦灵珍, 王佳, 孙新晓, 袁其朋, 申晓林. 黄酮类化合物生物合成及其在化妆品中应用的研究[J]. 合成生物学, 2025, 6(2): 373-390. |
| [9] | 肖森, 胡立涛, 石智诚, 王发银, 余思婷, 堵国成, 陈坚, 康振. 可控分子量透明质酸的生物合成研究进展[J]. 合成生物学, 2025, 6(2): 445-460. |
| [10] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [11] | 左一萌, 张姣姣, 连佳长. 酿酒酵母使能技术在化妆品原料合成中的应用[J]. 合成生物学, 2025, 6(2): 233-253. |
| [12] | 汤传根, 王璟, 张烁, 张昊宁, 康振. 功能肽合成和挖掘策略研究进展[J]. 合成生物学, 2025, 6(2): 461-478. |
| [13] | 郭婷婷, 韩湘凝, 黄熙婷, 张婷婷, 孔健. 乳酸菌的合成生物学工具及在合成益肤因子中的应用[J]. 合成生物学, 2025, 6(2): 320-333. |
| [14] | 张萍, 张维娇, 胥睿睿, 李江华, 陈坚, 康振. 防晒化合物类菌孢素氨基酸的生物合成[J]. 合成生物学, 2025, 6(2): 306-319. |
| [15] | 黄姝涵, 马赫, 罗云孜. 生物合成红景天苷的研究进展[J]. 合成生物学, 2025, 6(2): 391-407. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||