合成生物学 ›› 2022, Vol. 3 ›› Issue (3): 445-464.DOI: 10.12211/2096-8280.2022-013
分子伴侣作用下的蛋白质稳定与进化
唐宇琦1,2, 叶松涛1,2, 刘嘉1,2, 张鑫1,2
- 1.西湖大学理学院化学系,浙江 杭州 310030
2.浙江西湖高等研究院,理学研究所,浙江 杭州 310024
-
收稿日期:2022-02-12修回日期:2022-05-07出版日期:2022-06-30发布日期:2022-07-13 -
通讯作者:张鑫 -
作者简介:唐宇琦 (1998 —),女,科研助理。研究方向是对体外蛋白质相分离的理解。E-mail: tangyuqi@westlake.edu.cn张鑫 (1978 —),男,教授,博士生导师。张鑫 课题组聚焦于化学和生物的交叉领域,以“生物聚集体化学”为研究中心,瞄准此研究领域亟需解决的重要科学和技术问题,为基础研究和生物医药产业提供重要科学支持。E-mail: zhangxin@westlake.edu.cn
Molecular chaperones promote protein stability and evolution
TANG Yuqi1,2, YE Songtao1,2, LIU Jia1,2, ZHANG Xin1,2
- 1.Department of Chemistry,School of Science,Westlake University,Hangzhou 310030,Zhejiang,China
2.Institute of Natural Sciences,Westlake Institute for Advanced Study,Hangzhou 310024,Zhejiang,China
-
Received:2022-02-12Revised:2022-05-07Online:2022-06-30Published:2022-07-13 -
Contact:ZHANG Xin
摘要:
新生多肽链通常需要折叠成独特的三维结构来发挥其生物学功能。天然存在的蛋白质仅具有边缘稳定性,少量突变或轻微环境扰动就可能影响蛋白质的正确折叠。蛋白质组的稳定性,即蛋白质稳态,由蛋白质组中较不稳定的蛋白质决定,因而也具有边缘稳定性。蛋白质以及蛋白质组的边缘稳定性决定了细胞内存在着复杂的质量控制机制,用来帮助蛋白质正确折叠、修复或降解错误折叠的蛋白质。本文详细介绍了以热休克蛋白家族为代表的分子伴侣协助蛋白质折叠的内部机制,并回顾了通过过量表达分子伴侣、转录因子等手段提高蛋白质稳态的研究。蛋白质在保持稳定性的同时也在不断进化,本文介绍了蛋白质稳定性与可进化性关系的研究。实验证明,稳定性增强的蛋白质提高了对随机突变的包容度,有助于积累更多突变。相较于野生型蛋白质,这些蛋白质突变体在不同环境的选择下,会产生更多功能适应性突变体,即发生进化。因而蛋白质的稳定性是影响其进化的重要因素。分子伴侣作为蛋白质折叠的参与者,直接协助了蛋白质的定向进化。本文围绕蛋白质折叠的稳定性、蛋白质稳态和蛋白质进化的问题,讨论了以分子伴侣为主的分子机器帮助维持蛋白质稳定、促进蛋白质进化的相关研究。鉴于生物系统的复杂程度,我们对生物进化的理解仍然有限。希望关于影响蛋白质稳定性和可进化性的研究能够为理解蛋白质结构功能的构效关系提供独特见解,同时也为探究蛋白质相关疾病致病机理提供理论基础。
引用本文
唐宇琦, 叶松涛, 刘嘉, 张鑫. 分子伴侣作用下的蛋白质稳定与进化[J]. 合成生物学, 2022, 3(3): 445-464.
TANG Yuqi, YE Songtao, LIU Jia, ZHANG Xin. Molecular chaperones promote protein stability and evolution[J]. Synthetic Biology Journal, 2022, 3(3): 445-464.
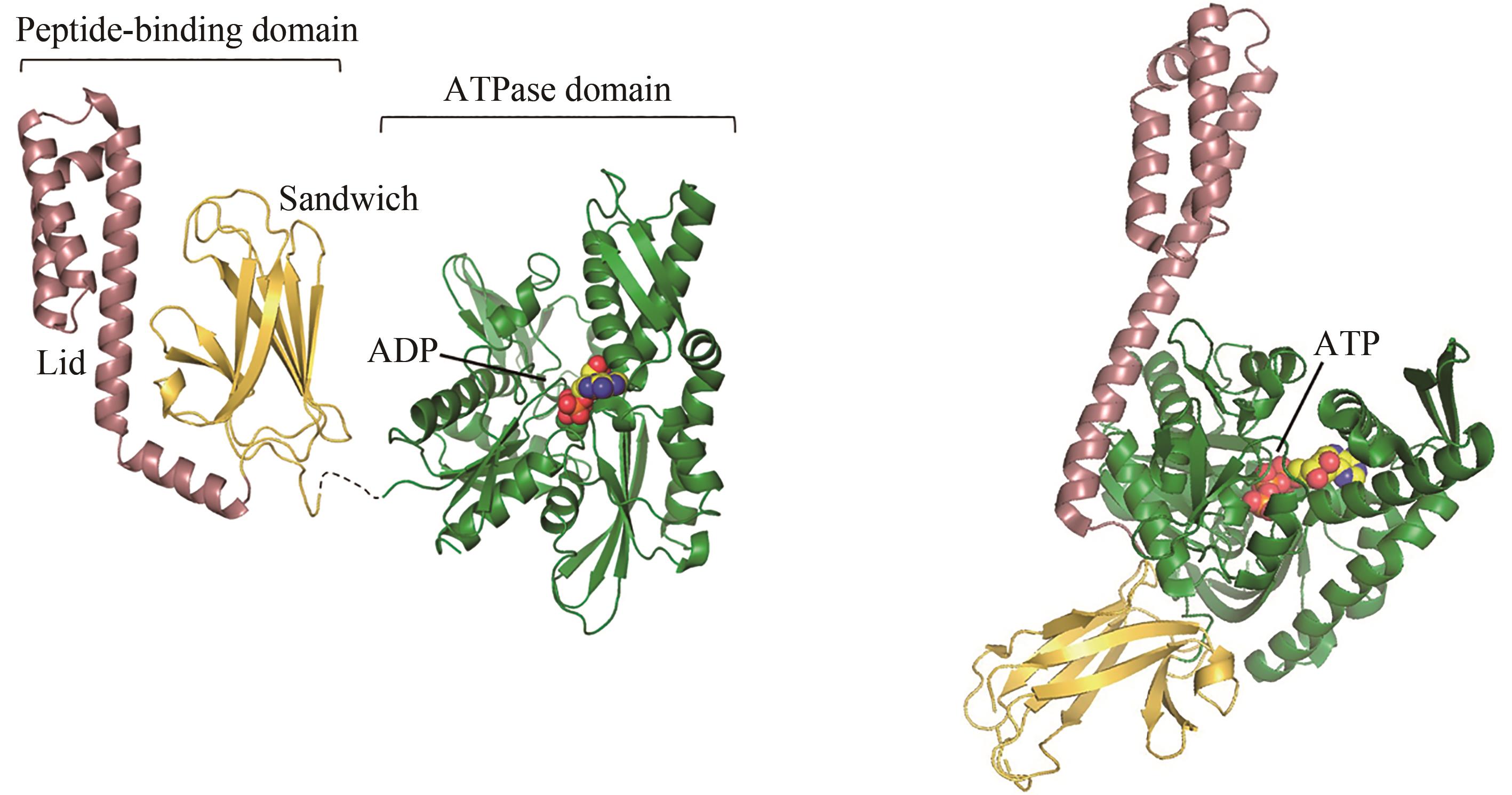
图2 ADP结合(左)和ATP结合(右,PDB代码:4B9Q)[55]的Hsp70构象[在ADP结合状态下,肽链结合结构域(PDB代码:1DKZ)[50]和 ATP酶结构域(PDB代码:3HSC)[51]通过灵活的氨基酸链连接]
Fig. 2 ADP-bound (left) and ATP-bound (right, PDB code: 4B9Q)[55] conformations of Hsp70[In the ADP-bound state, the peptide-binding domain (PDB code: 1DKZ)[50] and ATPase domain (PDB code: 3HSC)[51] are connected by a flexible linker]
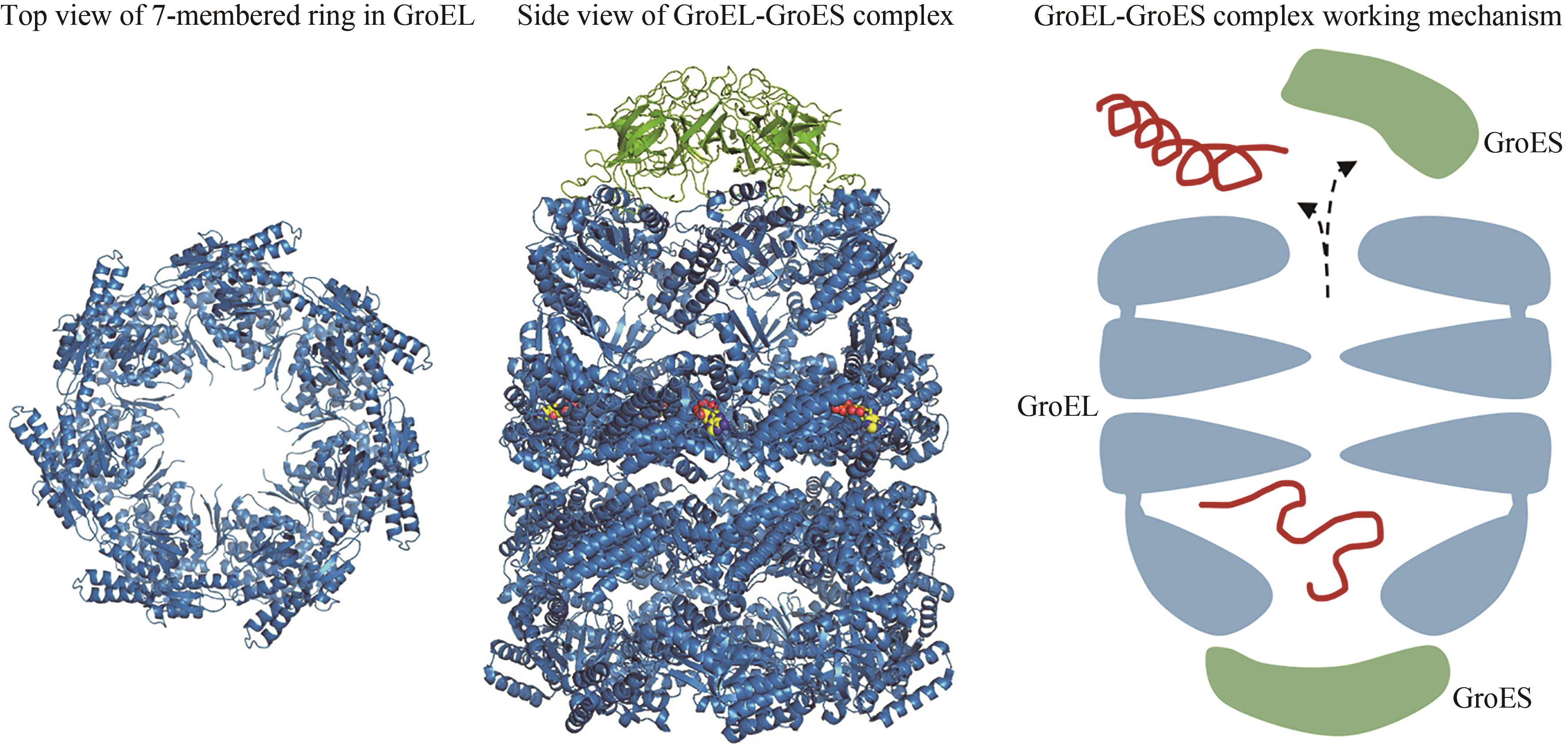
图3 Hsp60七元环(左,PDB代码:1IOK)[65]和Hsp60/Hsp10复合物(中,PDB代码:1AON)[67]的结构,以及Hsp60/Hsp10作用机理(右)
Fig. 3 Structures of the 7-membered ring in GroEL (left, PDB code: 1IOK)[65] and the Hsp60/Hsp10(middle, PDB code: 1AON)[67], as well as the working mechanism of Hsp60/Hsp10 (right)
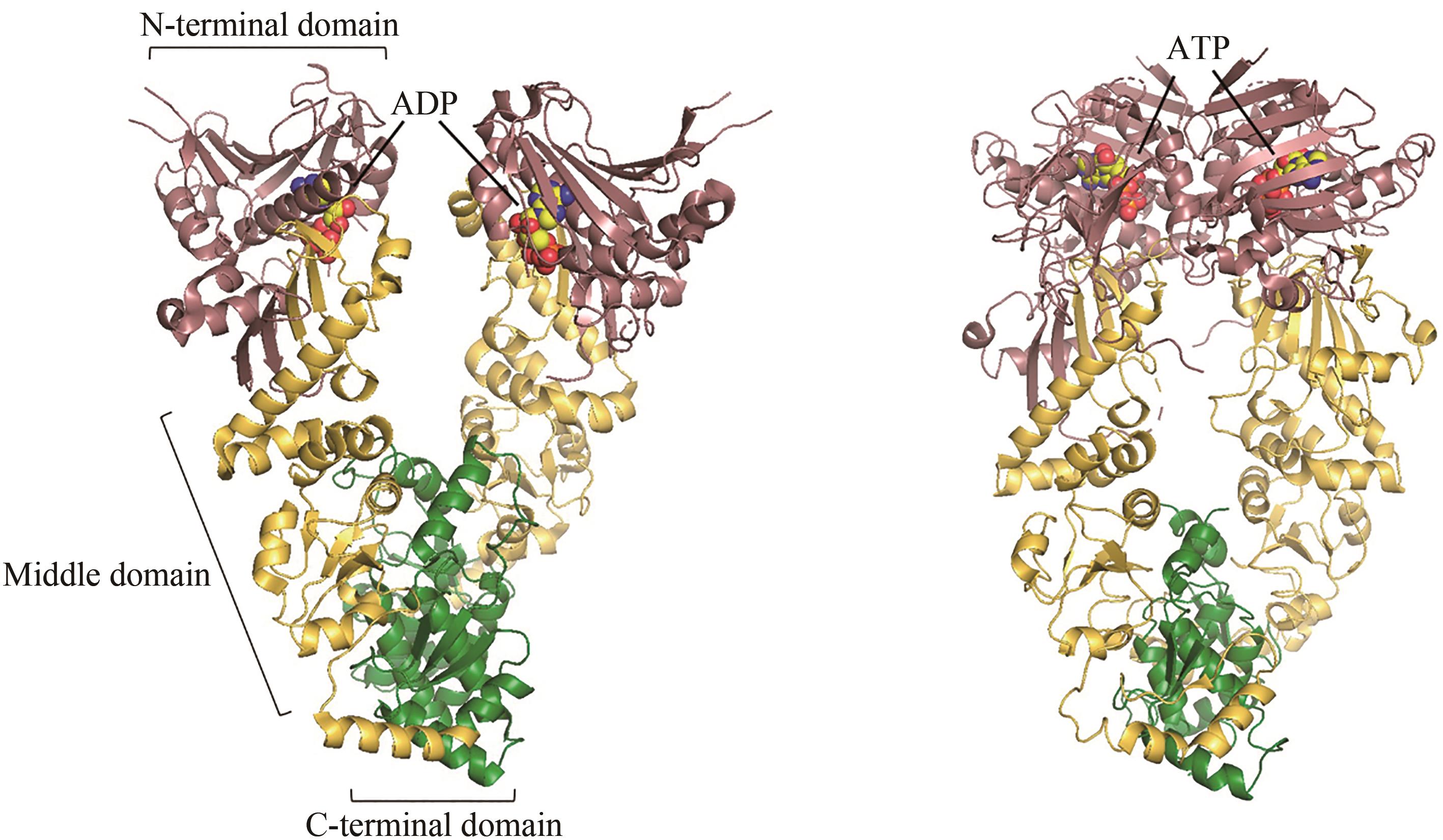
图4 ADP结合(左,PDB代码:2O1V)[79]和ATP结合(右,PDB代码:2CG9)[76]的Hsp90构象
Fig. 4 ADP-bound (left, PDB code: 2O1V)[79] and ATP-bound (right, PDB code: 2CG9)[76] conformations of Hsp90
| 1 | GIVER L, GERSHENSON A, FRESKGARD P O, et al. Directed evolution of a thermostable esterase[J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(22): 12809-12813. |
| 2 | VAN DEN BURG B, VRIEND G, VELTMAN O R, et al. Engineering an enzyme to resist boiling[J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(5): 2056-2060. |
| 3 | MIYAZAKI K, WINTRODE P L, GRAYLING R A, et al. Directed evolution study of temperature adaptation in a psychrophilic enzyme[J]. Journal of Molecular Biology, 2000, 297(4): 1015-1026. |
| 4 | GONZÁLEZ M, BAGATOLLI L A, ECHABE I, et al. Interaction of biotin with streptavidin: Thermostability and conformational changes upon binding[J]. Journal of Biological Chemistry, 1997, 272(17): 11288-11294. |
| 5 | SANCHEZ DE GROOT N, ARMAOS A, GRAÑA-MONTES R, et al. RNA structure drives interaction with proteins[J]. Nature Communications, 2019, 10: 32-46. |
| 6 | LAGIER-TOURENNE C, POLYMENIDOU M, CLEVELAND D W. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration[J]. Human Molecular Genetics, 2010, 19(R1): R46-R64. |
| 7 | KAPELI K, PRATT G A, VU A Q, et al. Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses[J]. Nature Communications, 2016, 7: 12143. |
| 8 | STRICKLER S S, GRIBENKO A V, GRIBENKO A V, et al. Protein stability and surface electrostatics: A charged relationship[J]. Biochemistry, 2006, 45(9): 2761-2766. |
| 9 | CURTIS R A, PRAUSNITZ J M, BLANCH H W. Protein-protein and protein-salt interactions in aqueous protein solutions containing concentrated electrolytes[J]. Biotechnology and Bioengineering, 1998, 57(1): 11-21. |
| 10 | LABER J R, DEAR B J, MARTINS M L, et al. Charge shielding prevents aggregation of supercharged GFP variants at high protein concentration[J]. Molecular Pharmaceutics, 2017, 14(10): 3269-3280. |
| 11 | TAVERNA D M, GOLDSTEIN R A. Why are proteins marginally stable? [J]. Proteins: Structure, Function, and Genetics, 2002, 46(1): 105-109. |
| 12 | VOGL T, JATZKE C, HINZ H J, et al. Thermodynamic stability of annexin V E17G: Equilibrium parameters from an irreversible unfolding reaction[J]. Biochemistry, 1997, 36(7): 1657-1668. |
| 13 | GHOSH K, DILL K. Cellular proteomes have broad distributions of protein stability[J]. Biophysical Journal, 2010, 99(12): 3996-4002. |
| 14 | ROSS C A, POIRIER M A. Protein aggregation and neurodegenerative disease[J]. Nature Medicine, 2004, 10(7): S10-S17. |
| 15 | AGUZZI A, O'CONNOR T. Protein aggregation diseases: Pathogenicity and therapeutic perspectives[J]. Nature Reviews Drug Discovery, 2010, 9(3): 237-248. |
| 16 | 陈冬冬, 石琦, 董小平. 蛋白质体外扩增技术在神经退行性疾病诊断中的应用[J]. 疾病监测, 2022, 37(2): 180-184. |
| CHEN D D, SHI Q, DONG X P. Application of protein amplification in vitro in the diagnosis of neurodegenerative diseases[J]. Disease Surveillance, 2022, 37(2): 180-184. | |
| 17 | 张肖楠, 廉姜芳. 内质网应激与蛋白质错误折叠相关疾病的研究进展[J]. 浙江医学, 2020, 42(1): 86-90. |
| ZHANG X N, LIAN J F. Research progress of endoplasmic reticulum stress and protein misfolding-related diseases [J]. Zhejiang Medical Journal, 2020, 42(1): 86-90. | |
| 18 | 熊秋宏, 李文静, 吴长新. 自噬和泛素-蛋白酶体系统之间的交联[J]. 中国生物化学与分子生物学报, 2018, 34(11): 1154-1159. |
| XIONG Q H, LI W J, WU C X. The crosstalk between autophagy and ubiquitin-proteasome system[J]. Chinese Journal of Biochemistry and Molecular Biology, 2018, 34(11): 1154-1159. | |
| 19 | OTTENS F, FRANZ A, HOPPE T. Build-UPS and break-downs: Metabolism impacts on proteostasis and aging[J]. Cell Death & Differentiation, 2021, 28(2): 505-521. |
| 20 | DONG Z, CUI H J. The autophagy-lysosomal pathways and their emerging roles in modulating proteostasis in tumors[J]. Cells, 2018, 8(1): 4. |
| 21 | HARTL F U, MARTIN J. Molecular chaperones in cellular protein folding[J]. Current Opinion in Structural Biology, 1995, 5(1): 92-102. |
| 22 | MORÁN LUENGO T, MAYER M P, RÜDIGER S G D. The Hsp70-Hsp90 chaperone cascade in protein folding[J]. Trends in Cell Biology, 2019, 29(2): 164-177. |
| 23 | HARTL F U, BRACHER A, HAYER-HARTL M. Molecular chaperones in protein folding and proteostasis[J]. Nature, 2011, 475(7356): 324-332. |
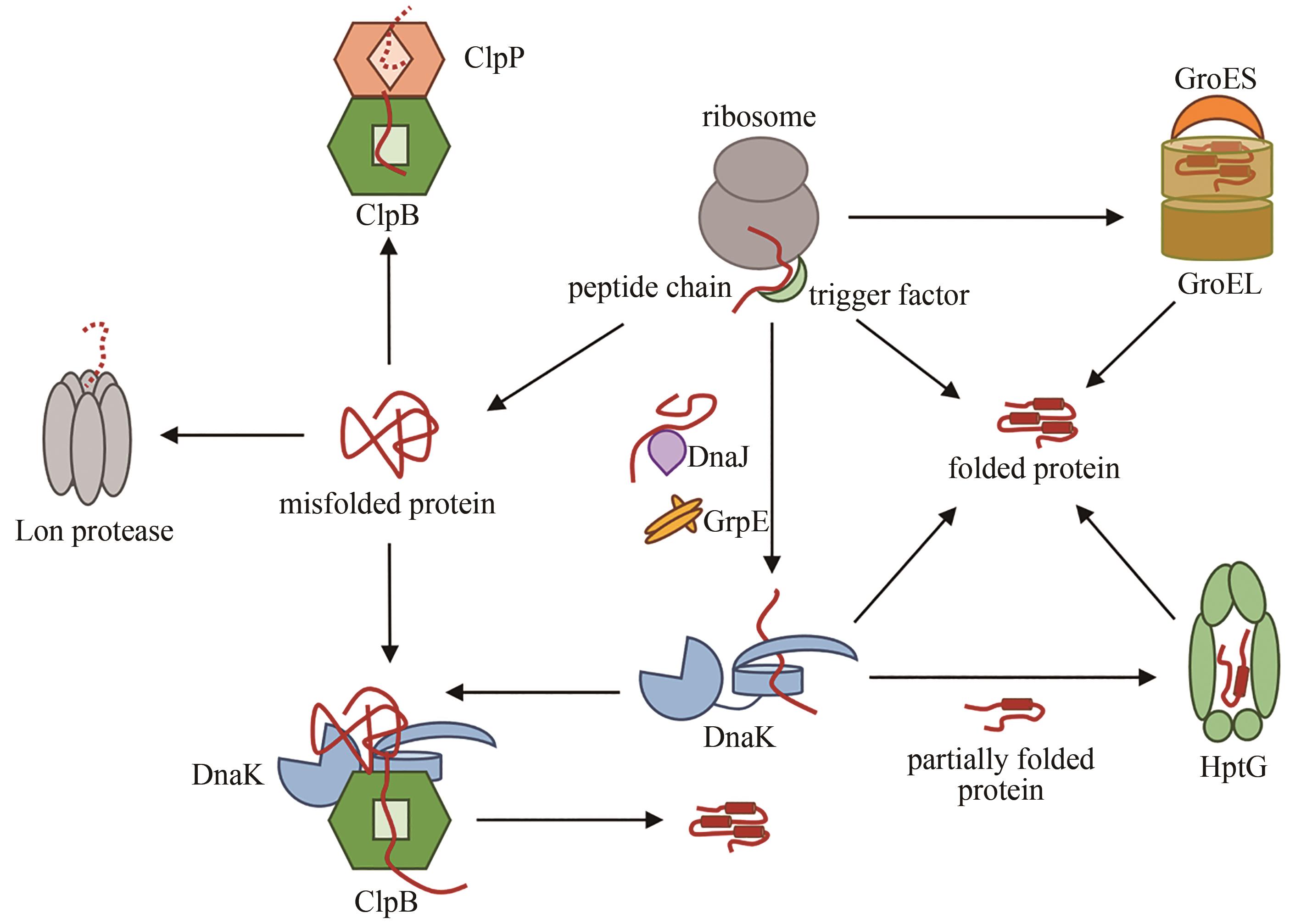
| 24 | KAISER C M, CHANG H C, AGASHE V R, et al. Real-time observation of trigger factor function on translating ribosomes[J]. Nature, 2006, 444(7118): 455-460. |
| 25 | DOYLE S M, GENEST O, WICKNER S. Protein rescue from aggregates by powerful molecular chaperone machines[J]. Nature Reviews Molecular Cell Biology, 2013, 14(10): 617-629. |
| 26 | TAVARIA M, GABRIELE T, KOLA I, et al. A hitchhiker's guide to the human Hsp70 family[J]. Cell Stress & Chaperones, 1996, 1(1): 23-28. |
| 27 | THOMAS J G, BANEYX F. Protein misfolding and inclusion body formation in recombinant escherichia coli cells overexpressing heat-shock proteins[J]. Journal of Biological Chemistry, 1996, 271(19): 11141-11147. |
| 28 | HALDAR S, TAPIA-ROJO R, ECKELS E C, et al. Trigger factor chaperone acts as a mechanical foldase[J]. Nature Communications, 2017, 8: 668. |
| 29 | NUNES J M, MAYER-HARTL M, HARTL F U, et al. Action of the Hsp70 chaperone system observed with single proteins[J]. Nature Communications, 2015, 6: 6307. |
| 30 | ASRAFUZZAMAN S, PATRA M, ROY S S, et al. The holding and folding chaperone properties of two small heat-shock pair proteins IbpA and IbPB of Escherichia coli [J]. Biophysical Journal, 2011, 100(3): 209a-210a. |
| 31 | DEVILLE C, FRANKE K, MOGK A, et al. Two-step activation mechanism of the ClpB disaggregase for sequential substrate threading by the main ATPase motor[J]. Cell Reports, 2019, 27(12): 3433-3446.e4. |
| 32 | 郑岩.不同分子伴侣活性对新生肽链的影响[D].哈尔滨: 哈尔滨工业大学,2020. |
| ZHENG Yan. Effects of different molecular chaperone activities on nascent peptide chains[D].Harbin: Harbin Institute of Technology,2020. | |
| 33 | RIZZOLO K, YU A Y H, OLOGBENLA A, et al. Functional cooperativity between the trigger factor chaperone and the ClpXP proteolytic complex[J]. Nature Communications, 2021, 12: 281. |
| 34 | WRUCK F, AVELLANEDA M J, KOERS E J, et al. Protein folding mediated by trigger factor and Hsp70: new insights from single-molecule approaches[J]. Journal of Molecular Biology, 2018, 430(4): 438-449. |
| 35 | BHANDARI V, HOURY W A. Substrate interaction networks of the Escherichia coli chaperones: Trigger factor, DnaK and GroEL[J]. Advances in Experimental Medicine and Biology, 2015, 883: 271-294. |
| 36 | AGASHE V R, GUHA S, CHANG H C, et al. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed[J]. Cell, 2004, 117(2): 199-209. |
| 37 | LASKOWSKA E, WAWRZYNÓW A, TAYLOR A. IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock[J]. Biochimie, 1996, 78(2): 117-122. |
| 38 | HASLBECK M, VIERLING E. A first line of stress defense: small heat shock proteins and their function in protein homeostasis[J]. Journal of Molecular Biology, 2015, 427(7): 1537-1548. |
| 39 | ALLEN S P, POLAZZI J O, GIERSE J K, et al. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli [J]. Journal of Bacteriology, 1992, 174(21): 6938-6947. |
| 40 | MOGK A, DEUERLING E, VORDERWÜLBECKE S, et al. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation[J]. Molecular Microbiology, 2003, 50(2): 585-595. |
| 41 | SCHOPF F H, BIEBL M M, BUCHNER J. The HSP90 chaperone machinery[J]. Nature Reviews Molecular Cell Biology, 2017, 18(6): 345-360. |
| 42 | SAIBIL H. Chaperone machines for protein folding, unfolding and disaggregation[J]. Nature Reviews Molecular Cell Biology, 2013, 14(10): 630-642. |
| 43 | CALLONI G, CHEN T T, SCHERMANN S M, et al. DnaK functions as a central hub in the E. coli chaperone network[J]. Cell Reports, 2012, 1(3): 251-264. |
| 44 | MISSELWITZ B, STAECK O, RAPOPORT T A. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences[J]. Molecular Cell, 1998, 2(5): 593-603. |
| 45 | JIANG Y J, ROSSI P, KALODIMOS C G. Structural basis for client recognition and activity of Hsp40 chaperones[J]. Science, 2019, 365(6459): 1313-1319. |
| 46 | SARBENG E B, LIU Q D, TIAN X L, et al. A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein[J]. Journal of Biological Chemistry, 2015, 290(14): 8849-8862. |
| 47 | LIU Q D, LI H T, YANG Y, et al. A disulfide-bonded DnaK dimer is maintained in an ATP-bound state[J]. Cell Stress & Chaperones, 2017, 22(2): 201-212. |
| 48 | MAYER M P. Gymnastics of molecular chaperones[J]. Molecular Cell, 2010, 39(3): 321-331. |
| 49 | KAMPINGA H H, CRAIG E A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity[J]. Nature Reviews Molecular Cell Biology, 2010, 11(8): 579-592. |
| 50 | ZHU X, ZHAO X, BURKHOLDER W F, et al. Structural analysis of substrate binding by the molecular chaperone DnaK[J]. Science, 1996, 272(5268): 1606-1614. |
| 51 | FLAHERTY K M, DELUCA-FLAHERTY C, MCKAY D B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein[J]. Nature, 1990, 346(6285): 623-628. |
| 52 | KITYK R, KOPP J, MAYER M P. Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones[J]. Molecular Cell, 2018, 69(2): 227-237. |
| 53 | YU H Y, ZIEGELHOFFER T, OSIPIUK J, et al. Roles of intramolecular and intermolecular interactions in functional regulation of the Hsp70 J-protein co-chaperone Sis1[J]. Journal of Molecular Biology, 2015, 427(7): 1632-1643. |
| 54 | YU H Y, ZIEGELHOFFER T, CRAIG E A. Functionality of class A and class B J-protein co-chaperones with Hsp70[J]. FEBS Letters, 2015, 589(19PartB): 2825-2830. |
| 55 | KITYK R, KOPP J, SINNING I, et al. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones[J]. Molecular Cell, 2012, 48(6): 863-874. |
| 56 | RÜDIGER S, BUCHBERGER A, BUKAU B. Interaction of Hsp70 chaperones with substrates[J]. Nature Structural Biology, 1997, 4(5): 342-349. |
| 57 | LACKIE R E, MACIEJEWSKI A, OSTAPCHENKO V G, et al. The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases[J]. Frontiers in Neuroscience, 2017, 11: 254. |
| 58 | MOGK A, KUMMER E, BUKAU B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation[J]. Frontiers in Molecular Biosciences, 2015, 2: 22. |
| 59 | CRAIG E A. Hsp70 at the membrane: Driving protein translocation[J]. BMC Biology, 2018, 16(1): 11. |
| 60 | SU P H, LI H M. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts[J]. The Plant Cell, 2010, 22(5): 1516-1531. |
| 61 | SAIBIL H R, FENTON W A, CLARE D K, et al. Structure and allostery of the chaperonin GroEL[J]. Journal of Molecular Biology, 2013, 425(9): 1476-1487. |
| 62 | HAYER-HARTL M, BRACHER A, HARTL F U. The GroEL-GroES chaperonin machine: A nano-cage for protein folding[J]. Trends in Biochemical Sciences, 2016, 41(1): 62-76. |
| 63 | CHEN D H, MADAN D M, WEAVER J, et al. Visualizing GroEL/ES in the act of encapsulating a folding protein[J]. Cell, 2013, 153(6): 1354-1365. |
| 64 | BRAIG K, OTWINOWSKI Z, HEGDE R, et al. The crystal structure of the bacterial chaperonin GroEL at 2.8 A[J]. Nature, 1994, 371(6498): 578-586. |
| 65 | FUKAMI T A, YOHDA M, TAGUCHI H, et al. Crystal structure of chaperonin-60 from paracoccus denitrificans[J]. Journal of Molecular Biology, 2001, 312(3): 501-509. |
| 66 | FIAUX J, BERTELSEN E B, HORWICH A L, et al. NMR analysis of a 900K GroEL-GroES complex[J]. Nature, 2002, 418(6894): 207-211. |
| 67 | XU Z H, HORWICH A L, SIGLER P B. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex[J]. Nature, 1997, 388(6644): 741-750. |
| 68 | HORWICH A L, FENTON W A. Chaperonin-mediated protein folding: Using a central cavity to kinetically assist polypeptide chain folding[J]. Quarterly Reviews of Biophysics, 2009, 42(2): 83-116. |
| 69 | XU Z H, HORWICH A L, SIGLER P B. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex[J]. Nature, 1997, 388(6644): 741-750. |
| 70 | HYEON C, LORIMER G H, THIRUMALAI D. Dynamics of allosteric transitions in GroEL[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(50): 18939-18944. |
| 71 | CARUSO BAVISOTTO C, ALBERTI G, VITALE A M, et al. Hsp60 post-translational modifications: Functional and pathological consequences[J]. Frontiers in Molecular Biosciences, 2020, 7: 95. |
| 72 | WHITESELL L, LINDQUIST S L. HSP90 and the chaperoning of cancer[J]. Nature Reviews Cancer, 2005, 5(10): 761-772. |
| 73 | LUO W J, SUN W L, TALDONE T, et al. Heat shock protein 90 in neurodegenerative diseases[J]. Molecular Neurodegeneration, 2010, 5: 24. |
| 74 | CSERMELY P, SCHNAIDER T, SŐTI C, et al. The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review[J]. Pharmacology & Therapeutics, 1998, 79(2): 129-168. |
| 75 | DIDENKO T, DUARTE A M S, KARAGÖZ G E, et al. Hsp90 structure and function studied by NMR spectroscopy[J]. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2012, 1823(3): 636-647. |
| 76 | ALI M M U, ROE S M, VAUGHAN C K, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex[J]. Nature, 2006, 440(7087): 1013-1017. |
| 77 | PRODROMOU C. Mechanisms of Hsp90 regulation[J]. The Biochemical Journal, 2016, 473(16): 2439-2452. |
| 78 | KARAGÖZ G E, RÜDIGER S G D. Hsp90 interaction with clients[J]. Trends in Biochemical Sciences, 2015, 40(2): 117-125. |
| 79 | DOLLINS D E, WARREN J J, IMMORMINO R M, et al. Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones[J]. Molecular Cell, 2007, 28(1): 41-56. |
| 80 | LI J, RICHTER K, REINSTEIN J, et al. Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle[J]. Nature Structural & Molecular Biology, 2013, 20(3): 326-331. |
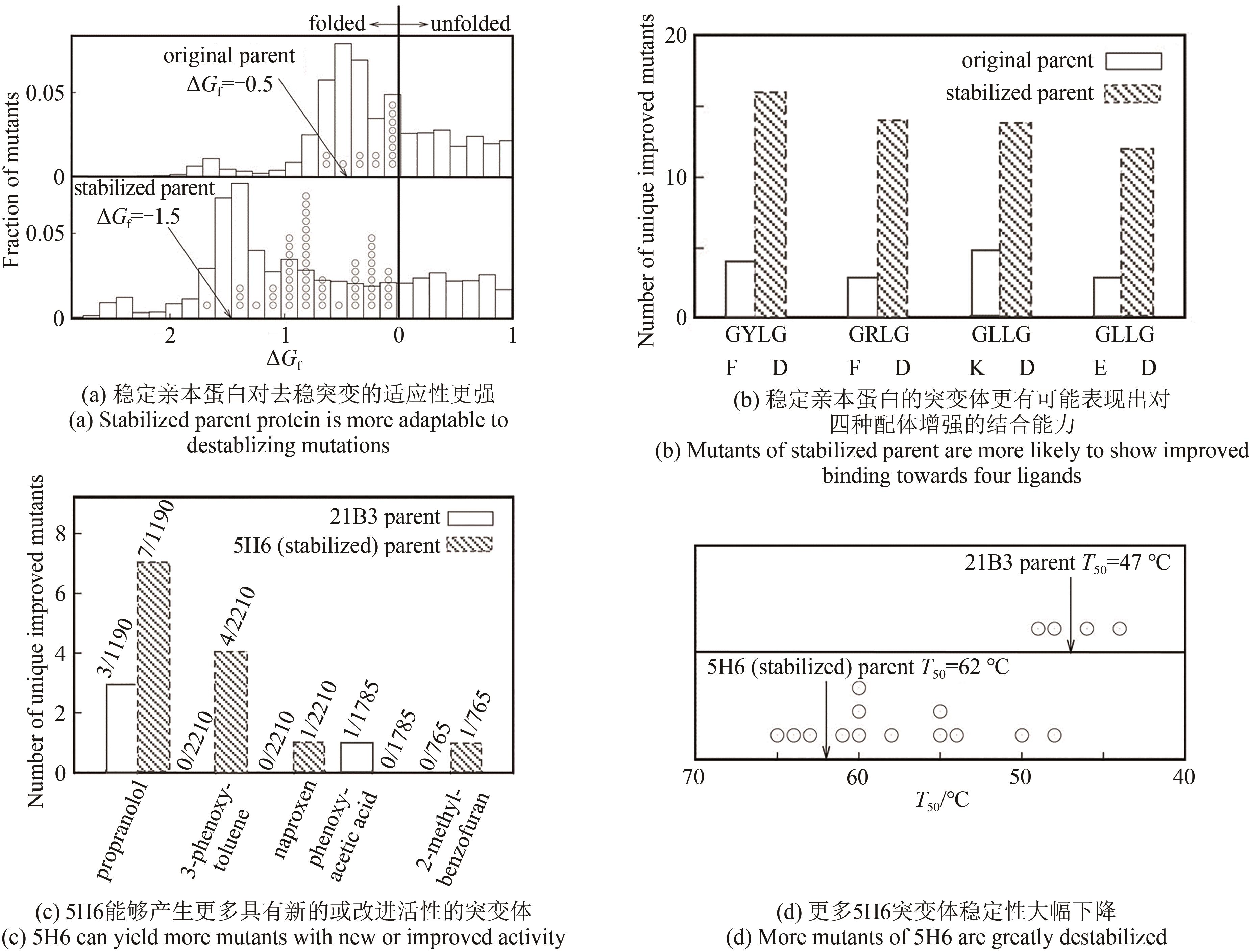
| 81 | VERBA K A, WANG R Y R, ARAKAWA A, et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase[J]. Science, 2016, 352(6293): 1542-1547. |
| 82 | WU F H, YUAN Y, LI D, et al. Extracellular HSPA1A promotes the growth of hepatocarcinoma by augmenting tumor cell proliferation and apoptosis-resistance[J]. Cancer Letters, 2012, 317(2): 157-164. |
| 83 | JAGADISH N, PARASHAR D, GUPTA N, et al. Heat shock protein 70-2 (HSP70-2) is a novel therapeutic target for colorectal cancer and is associated with tumor growth[J]. BMC Cancer, 2016, 16: 561. |
| 84 | KAMPINGA H H, HAGEMAN J, VOS M J, et al. Guidelines for the nomenclature of the human heat shock proteins[J]. Cell Stress & Chaperones, 2009, 14(1): 105-111. |
| 85 | CHEETHAM M E, CAPLAN A J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function[J]. Cell Stress & Chaperones, 1998, 3(1): 28-36. |
| 86 | VILASI S, et al. Chaperonin of group I: oligomeric spectrum and biochemical and biological implications.Frontiers in Mole cular Biosciences, 2017, 4: 99. |
| 87 | KUBOTA H, HYNES G, WILLISON K. The chaperonin containing t-complex polypeptide 1 (TCP-1). Multisubunit machinery assisting in protein folding and assembly in the eukaryotic cytosol[J]. European Journal of Biochemistry, 1995, 230(1): 3-16. |
| 88 | HOTER A, EL-SABBAN M E, NAIM H Y. The HSP90 family: structure, regulation, function, and implications in health and disease[J]. International Journal of Molecular Sciences, 2018, 19(9): 2560. |
| 89 | LEE S C, CHOI Y C, YU M H. Effect of the N-terminal hydrophobic sequence of hepatitis B virus surface antigen on the folding and assembly of hybrid beta-galactosidase in Escherichia coli [J]. European Journal of Biochemistry, 1990, 187(2): 417-424. |
| 90 | ZHANG X, LIU Y, GENEREUX J C, et al. Heat-shock response transcriptional program enables high-yield and high-quality recombinant protein production in Escherichia coli [J]. ACS Chemical Biology, 2014, 9(9): 1945-1949. |
| 91 | ZHAO K, LIU M Z, BURGESS R R. The global transcriptional response of Escherichia coli to induced sigma 32 protein involves sigma 32 regulon activation followed by inactivation and degradation of sigma 32 in vivo [J]. The Journal of Biological Chemistry, 2005, 280(18): 17758-17768. |
| 92 | GUISBERT E, HERMAN C, LU C Z, et al. A chaperone network controls the heat shock response in E. coli [J]. Genes & Development, 2004, 18(22): 2812-2821. |
| 93 | YURA T, GUISBERT E, PORITZ M, et al. Analysis of a 32 mutants defective in chaperone-mediated feedback control reveals unexpected complexity of the heat shock response[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(45): 17638-17643. |
| 94 | SUBBARAO SREEDHAR A, KALMÁR É, CSERMELY P, et al. Hsp90 isoforms: Functions, expression and clinical importance[J]. FEBS Letters, 2004, 562(1/2/3): 11-15. |
| 95 | CHEN B, PIEL W H, GUI L M, et al. The HSP90 family of genes in the human genome: insights into their divergence and evolution[J]. Genomics, 2005, 86(6): 627-637. |
| 96 | YUNO A, LEE M J, LEE S M, et al. Clinical evaluation and biomarker profiling of Hsp90 inhibitors[J]. Methods in Molecular Biology (Clifton, N J), 2018, 1709: 423-441. |
| 97 | NECKERS L, BLAGG B, HAYSTEAD T, et al. Methods to validate Hsp90 inhibitor specificity, to identify off-target effects, and to rethink approaches for further clinical development[J]. Cell Stress & Chaperones, 2018, 23(4): 467-482. |
| 98 | XIAO Y, LIU Y J. Recent advances in the discovery of novel HSP90 inhibitors: An update from 2014[J]. Current Drug Targets, 2020, 21(3): 302-317. |
| 99 | DAI C K, SAMPSON S B. HSF1: guardian of proteostasis in cancer[J]. Trends in Cell Biology, 2016, 26(1): 17-28. |
| 100 | YOON Y J, KIM J A, SHIN K D, et al. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter[J]. Journal of Biological Chemistry, 2011, 286(3): 1737-1747. |
| 101 | SALAMANCA H H, ANTONYAK M A, CERIONE R A, et al. Inhibiting heat shock factor 1 in human cancer cells with a potent RNA aptamer[J]. PLoS One, 2014, 9(5): e96330. |
| 102 | VILABOA N, BORÉ A, MARTIN-SAAVEDRA F, et al. New inhibitor targeting human transcription factor HSF1: effects on the heat shock response and tumor cell survival[J]. Nucleic Acids Research, 2017, 45(10): 5797-5817. |
| 103 | HUBEL P, URBAN C, BERGANT V, et al. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape[J]. Nature Immunology, 2019, 20(4): 493-502. |
| 104 | MA J, ZHANG X F, FENG Y B, et al. Structural and functional study of apoptosis-linked gene-2·Heme-binding protein 2 interactions in HIV-1 production[J]. Journal of Biological Chemistry, 2016, 291(52): 26670-26685. |
| 105 | MCKAY T B, HJORTDAL J, SEJERSEN H, et al. Differential effects of hormones on cellular metabolism in keratoconus in vitro [J]. Scientific Reports, 2017, 7: 42896. |
| 106 | CICATIELLO A G, DI GIROLAMO D, DENTICE M. Metabolic effects of the intracellular regulation of thyroid hormone: Old players, new concepts[J]. Frontiers in Endocrinology, 2018, 9: 474. |
| 107 | BRUICE T C, BENKOVIC S J. Chemical basis for enzyme catalysis[J]. Biochemistry, 2000, 39(21): 6267-6274. |
| 108 | AQVIST J, et al. Entropy and Enzyme Catalysis.Accounts of Chemical Reserc, 2017, 50(2): 199-207. |
| 109 | DRUMMOND D A, BLOOM J D, ADAMI C, et al. Why highly expressed proteins evolve slowly[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(40): 14338-14343. |
| 110 | BLOOM J D, WILKE C O, ARNOLD F H, et al. Stability and the evolvability of function in a model protein[J]. Biophysical Journal, 2004, 86(5): 2758-2764. |
| 111 | BLOOM J D, LABTHAVIKUL S T, OTEY C R, et al. Protein stability promotes evolvability[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(15): 5869-5874. |
| 112 | ZHENG J, GUO N, WAGNER A. Selection enhances protein evolvability by increasing mutational robustness and foldability[J]. Science, 2020, 370(6521): eabb5962. |
| 113 | JAMES L C, TAWFIK D S. Conformational diversity and protein evolution-a 60-year-old hypothesis revisited[J]. Trends in Biochemical Sciences, 2003, 28(7): 361-368. |
| 114 | TOKURIKI N, TAWFIK D S. Protein dynamism and evolvability[J]. Science, 2009, 324(5924): 203-207. |
| 115 | CREAN R M, GARDNER J M, KAMERLIN S C L. Harnessing conformational plasticity to generate designer enzymes[J]. Journal of the American Chemical Society, 2020, 142(26): 11324-11342. |
| 116 | QU, G, et al. Unlocking the stereoselectivity and substrate acceptance of enzymes: proline-induced loop engineering test[J]. Angewandte Chemie Internation Edition, 2022, 61(1): e202110793. |
| 117 | TOKURIKI N, TAWFIK D S. Chaperonin overexpression promotes genetic variation and enzyme evolution[J]. Nature, 2009, 459(7247): 668-673. |
| 118 | PHILLIPS A M, PONOMARENKO A I, CHEN K, et al. Destabilized adaptive influenza variants critical for innate immune system escape are potentiated by host chaperones[J]. PLoS Biology, 2018, 16(9): e3000008. |
| 119 | PHILLIPS A M, DOUD M B, GONZALEZ L O, et al. Enhanced ER proteostasis and temperature differentially impact the mutational tolerance of influenza hemagglutinin[J]. eLife, 2018, 7: e38795. |
| 120 | BANDYOPADHYAY A, SAXENA K, KASTURIA N, et al. Chemical chaperones assist intracellular folding to buffer mutational variations[J]. Nature Chemical Biology, 2012, 8(3): 238-245. |
| 121 | VELASQUEZ M T, RAMEZANI A, MANAL A, et al. Trimethylamine N-oxide: The good, the bad and the unknown[J]. Toxins, 2016, 8(11): 326. |
| 122 | QIAN Y Q, PATEL D, HARTL F U, et al. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain[J]. Journal of Molecular Biology, 1996, 260(2): 224-235. |
| 123 | CAPLAN A J, CYR D M, DOUGLAS M G. Eukaryotic homologues of Escherichia coli dnaJ: A diverse protein family that functions with hsp70 stress proteins[J]. Molecular Biology of the Cell, 1993, 4(6): 555-563. |
| 124 | ACHARYA P, KUMAR R, TATU U. Chaperoning a cellular upheaval in malaria: heat shock proteins in Plasmodium falciparum [J]. Molecular and Biochemical Parasitology, 2007, 153(2): 85-94. |
| 125 | FELDHEIM D, ROTHBLATT J, SCHEKMAN R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation[J]. Molecular and Cellular Biology, 1992, 12(7): 3288-3296. |
| 126 | SCHLENSTEDT G, HARRIS S, RISSE B, et al. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s[J]. The Journal of Cell Biology, 1995, 129(4): 979-988. |
| 127 | NISHIKAWA S, ENDO T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion[J]. The Journal of Biological Chemistry, 1997, 272(20): 12889-12892. |
| 128 | WANG J D, HERMAN C, TIPTON K A, et al. Directed evolution of substrate-optimized GroEL/S chaperonins[J]. Cell, 2002, 111(7): 1027-1039. |
| 129 | JACKREL M E, SHORTER J. Engineering enhanced protein disaggregases for neurodegenerative disease[J]. Prion, 2015, 9(2): 90-109. |
| [1] | 付雨, 钟芳锐. 化学原理驱动的光生物不对称催化研究进展[J]. 合成生物学, 2024, 5(5): 1021-1049. |
| [2] | 汤志军, 胡友财, 刘文. 酶促4+2和2+2环加成反应:区域与立体选择性的理解与应用[J]. 合成生物学, 2024, 5(3): 401-407. |
| [3] | 孙梦楚, 陆亮宇, 申晓林, 孙新晓, 王佳, 袁其朋. 基于荧光检测的高通量筛选技术和装备助力细胞工厂构建[J]. 合成生物学, 2023, 4(5): 947-965. |
| [4] | 明阳, 陈彬, 黄小强. 光酶催化合成进展[J]. 合成生物学, 2023, 4(4): 651-675. |
| [5] | 康里奇, 谈攀, 洪亮. 人工智能时代下的酶工程[J]. 合成生物学, 2023, 4(3): 524-534. |
| [6] | 阮青云, 黄莘, 孟子钧, 全舒. 蛋白质稳定性计算设计与定向进化前沿工具[J]. 合成生物学, 2023, 4(1): 5-29. |
| [7] | 祁延萍, 朱晋, 张凯, 刘彤, 王雅婕. 定向进化在蛋白质工程中的应用研究进展[J]. 合成生物学, 2022, 3(6): 1081-1108. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||