合成生物学 ›› 2022, Vol. 3 ›› Issue (6): 1201-1217.DOI: 10.12211/2096-8280.2022-041
梭菌分子遗传改造工具研究进展
刘佳昕1, 程驰1,2, 李欣启1, 汪超俊2, 张颖2, 薛闯1,2
- 1.大连理工大学生物工程学院,大连市合成生物学应用转化工程技术研究中心,辽宁 大连 116024
2.大连理工大学宁波研究院,浙江 宁波 315016
-
收稿日期:2022-07-18修回日期:2022-08-30出版日期:2022-12-31发布日期:2023-01-17 -
通讯作者:程驰,薛闯 -
作者简介:刘佳昕 (1998—),女,硕士研究生。主要进行生物学研究。E-mail:ljx123456@mail.dlut.edu.cn程驰 (1990—),女,副教授,研究生导师。主要研究领域为:①能源微生物遗传操作工具开发及代谢改造;②化学-生物耦合的CO2固定。E-mail:cheng.chi@dlut.edu.cn薛闯 (1982—),男,教授,博士生导师。主要从事生物质新能源的生产、分离纯化以及高效代谢菌株构建的研究:①生物法生产燃料乙醇及酵母菌的代谢网络研究;②微生物发酵法生产丁醇;③生物基化学品的高效分离;④有机膜的制备及分离技术;⑤先进能源生产菌株的构建及代谢途径信号转导;⑥激酶的生物信息分析。E-mail:xue.1@dlut.edu.cn -
基金资助:国家重点研发计划(2021YFC2102500);国家自然科学基金(21878035);大连市杰出青年科技人才支持计划(2021RJ03);大连理工大学基本科研业务费(DUT21RC(3)003, DUT22ZD102);辽宁省“百千万人才工程”经费
Recent progress in the molecular genetic modification tools of Clostridium
LIU Jiaxin1, CHENG Chi1,2, LI Xinqi1, WANG Chaojun2, ZHANG Ying2, XUE Chuang1,2
- 1.Engineering Research Center of Application and Transformation for Synthetic Biology Dalian,School of Bioengineering,Dalian University of Technology,Dalian 116024,Liaoning,China
2.NingBo Institute of Dalian University of Technology,Ningbo 315016,Zhejiang,China
-
Received:2022-07-18Revised:2022-08-30Online:2022-12-31Published:2023-01-17 -
Contact:CHENG Chi, XUE Chuang
摘要:
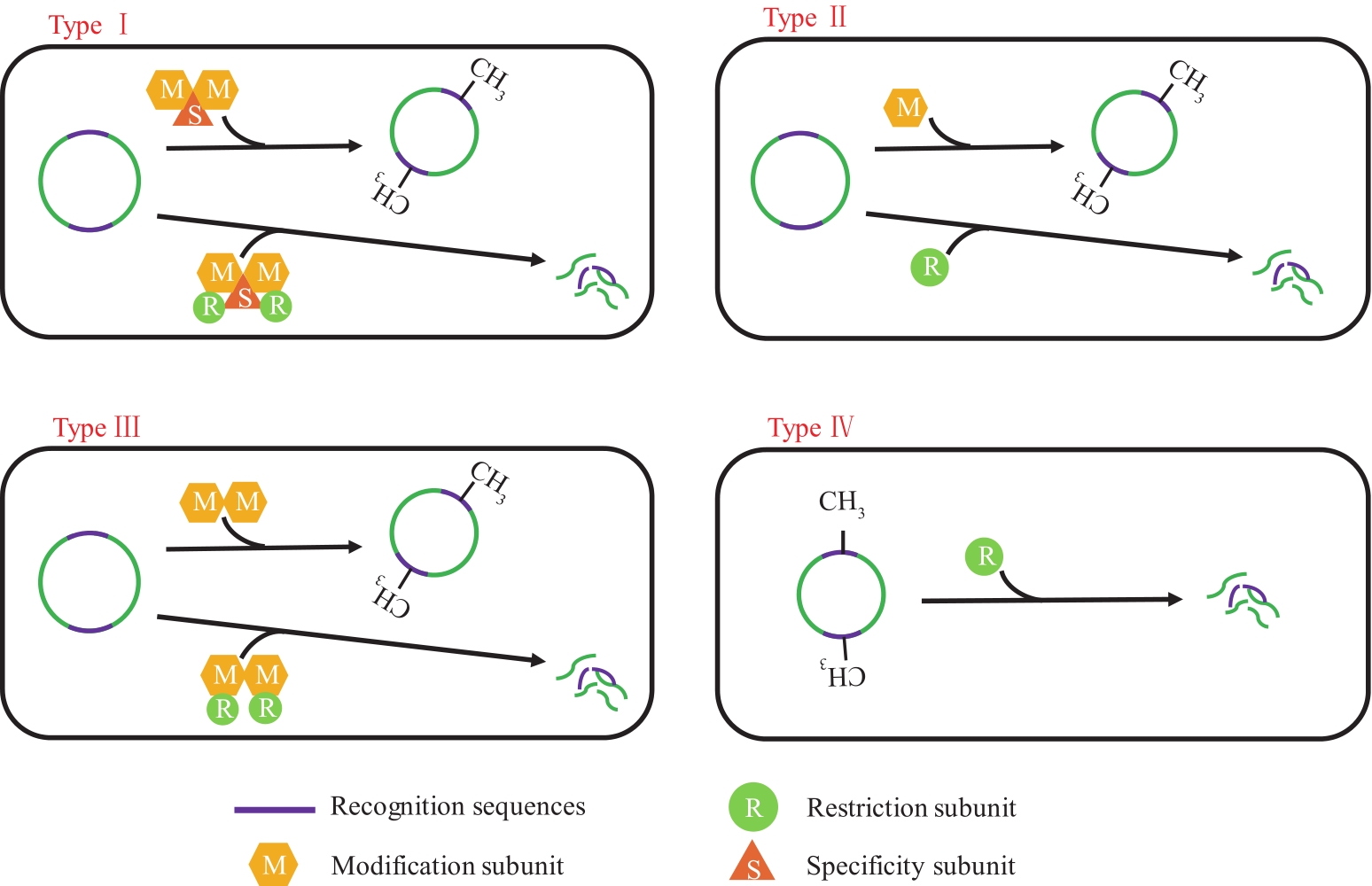
梭状芽孢杆菌是一类革兰氏阳性、可内生孢子的严格厌氧型细菌,可产生多种化学物质,包括现如今极具潜力的新型生物燃料丁醇。通过分子改造以提高梭菌发酵的浓度及产率一直是一项亟需突破的重要课题,但该方向的研究长期受限于梭菌不完善的遗传操作工具。近年来,随着分子生物学的快速发展,适用于梭菌的基因编辑工具不断发展,梭菌中已有反义RNA技术、TargeTron、基于同源重组或CRISPR/Cas系统介导的基因编辑技术等多种遗传操作工具,可以基本实现靶标基因插入、删除、替换、点突变以及表达水平调控等各种操作。文中对上述遗传操作工具研究进展进行了总结,并着重讨论了以重组酶为代表的新型遗传操作技术及其在梭菌中的应用潜力。今后应进一步优化现有的梭菌分子遗传改造工具,重点突破梭菌自身同源重组效率低下等技术难点,同时应大力发展新的基因编辑技术,如以CRISPR技术为核心的多位点共编辑系统、噬菌体重组酶介导的多拷贝定点和随机整合技术等。
引用本文
刘佳昕, 程驰, 李欣启, 汪超俊, 张颖, 薛闯. 梭菌分子遗传改造工具研究进展[J]. 合成生物学, 2022, 3(6): 1201-1217.
LIU Jiaxin, CHENG Chi, LI Xinqi, WANG Chaojun, ZHANG Ying, XUE Chuang. Recent progress in the molecular genetic modification tools of Clostridium[J]. Synthetic Biology Journal, 2022, 3(6): 1201-1217.
| 遗传操作工具 | 优点 | 缺点 | 梭菌菌株 | 基因 | 文献 |
|---|---|---|---|---|---|
| 基于游离质粒的基因过表达 | 设计简单,易于操作 | 质粒不稳定,需靠抗生素维持 | C. acetobutylicum ATCC 824 C. acetobutylicum DSM 792 C. paraputrificum M-21 C. tyrobutyricum JM1 C. tyrobutyricum ATCC 25755 C. perfringens SM101 C. cellulolyticum H10 | spo0A, hydA, adhE2, adC, groESL, txeR, tcdC, pdc-adhII | [ |
| 反义RNA技术 | 致死率低,易于筛选 | 仅在转录水平调控 | C. acetobutylicum ATCC 824 | ptb, buk, CoAT | [ |
| 转座子突变技术 | 可用于建立突变体库 | 难以控制插入位点 | C. difficile CD37 C. perfringens Strain 13 | random | [ |
| 二型内含子技术 | 操作简单,几乎适用于所有梭菌 | 脱靶概率较大,存在极性效应 | C. acetobutylicum ATCC 824 C. beijerinckii NCIMB 8052 | glcG, cbei2385, xylR, bdhA, bdhB, ptb, ack, adc | [ |
| 同源重组 | 可精确进行基因编辑 | 同源重组效率很低 | C. acetobutylicum NCIMB 8052 | gutD, spo0A | [ |
| 反筛标记介导的等位基因替换 | 相比纯粹的等位基因替换,效率有所提升 | 受制于第一次单交换效率 | C. acetobutylicum ATCC 824 | adh | [ |
| Ⅰ-SceⅠ归巢内切酶介导的等位基因替换 | 适用范围广,在革兰氏阳性和阴性菌中均可使用 | 设计复杂,操作烦琐,周期长 | C. acetobutylicum ATCC 824 C. beijerinckii NCIMB 8052 | adc, glcG, xylR | [ |
| 噬菌体丝氨酸重组酶介导的位点特异性基因编辑技术 | 适用于大片段DNA快速整合到宿主染色体上 | 受限于附着位点attb/p,应用范围小 | C. ljungdahlii DSM 13528 | thl, crt, bcd, etfB, etfA, hbd, ptb, buk | [ |
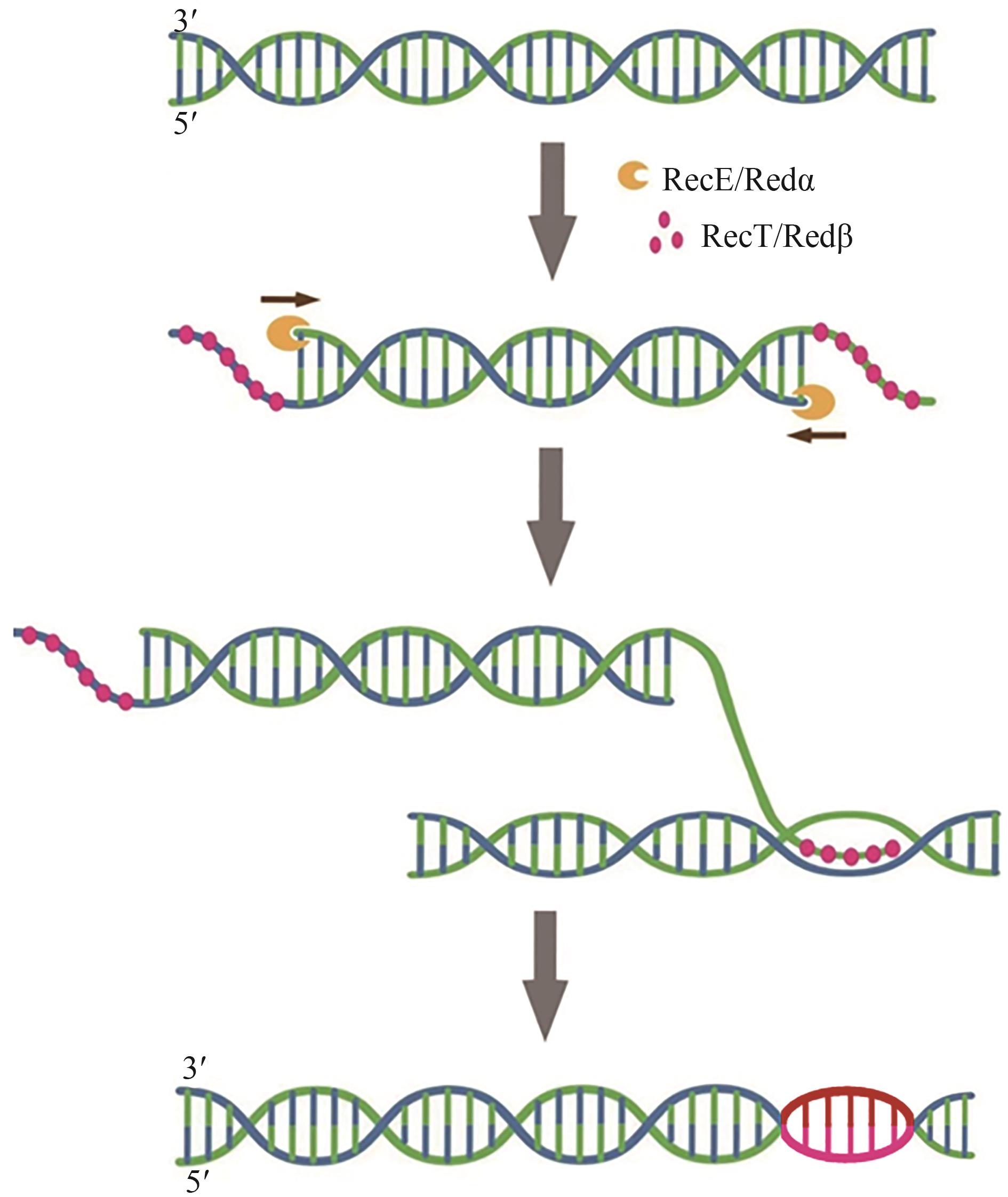
| Red/ET重组酶介导的同源重组 | 所需同源臂较短、不受限制性酶切位点限制 | 尚不完善 | C. acetobutylicum SMB009 | ermC | [ |
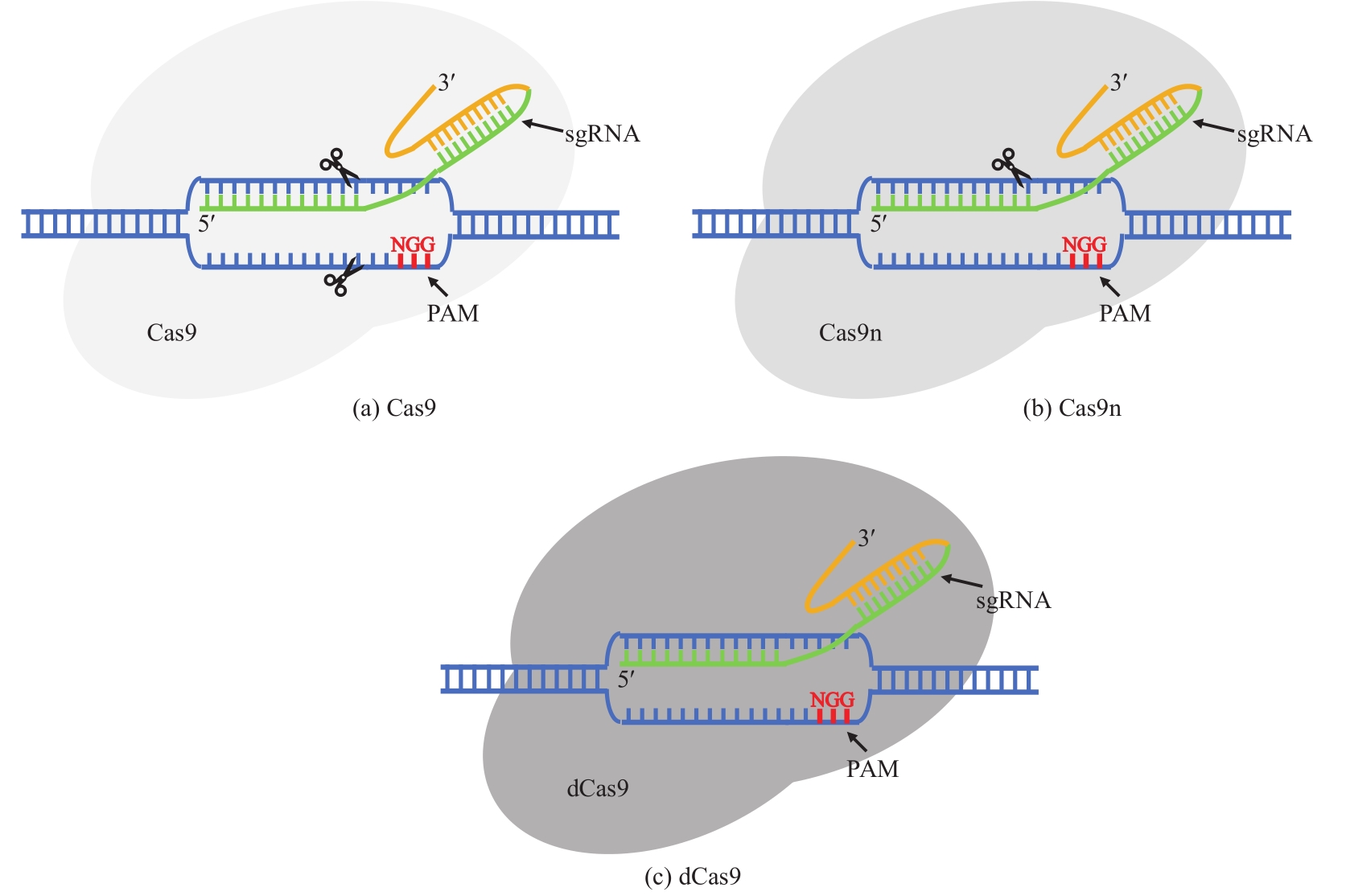
| CRISPR/Cas9系统介导的基因编辑 | 提高同源重组效率,可实现精确编辑 | 毒性大,很难获得转化子 | C. acetobutylicum DSM792 C. saccharoperbutylacetonicum N1-4 C. cellulovorans C. autoethanogenum DSM10061 | hprK, cac1502, pta, buk, clocel2243, adhE1, ctf,pyrE, 2,3-bdh | [ |
| CRISPR/Cas9n系统介导的基因编辑 | 相比于CRISPR/Cas9毒性有所降低,转化子数目增加 | 仍有毒性,转化子依然偏少 | C. cellulolyticum H10 C. acetobutylicum ATCC 824 C. beijerinckii NCIMB 8052 | pyrF, spo0A | [ |
| CRISPR/dCas9系统介导的基因表达下调 | 相比于CRISPR/Cas9毒性大大降低,易于得到转化子 | 依赖于sgRNA和基因;仅在转录水平调控 | C. pasteurianum ATCC 6013 C. acetobutylicum DSM792 | hprK, glpX | [ |
表1 梭菌遗传操作工具比较
Tab. 1 Comparison of different genetic tools applicable in Clostridium
| 遗传操作工具 | 优点 | 缺点 | 梭菌菌株 | 基因 | 文献 |
|---|---|---|---|---|---|
| 基于游离质粒的基因过表达 | 设计简单,易于操作 | 质粒不稳定,需靠抗生素维持 | C. acetobutylicum ATCC 824 C. acetobutylicum DSM 792 C. paraputrificum M-21 C. tyrobutyricum JM1 C. tyrobutyricum ATCC 25755 C. perfringens SM101 C. cellulolyticum H10 | spo0A, hydA, adhE2, adC, groESL, txeR, tcdC, pdc-adhII | [ |
| 反义RNA技术 | 致死率低,易于筛选 | 仅在转录水平调控 | C. acetobutylicum ATCC 824 | ptb, buk, CoAT | [ |
| 转座子突变技术 | 可用于建立突变体库 | 难以控制插入位点 | C. difficile CD37 C. perfringens Strain 13 | random | [ |
| 二型内含子技术 | 操作简单,几乎适用于所有梭菌 | 脱靶概率较大,存在极性效应 | C. acetobutylicum ATCC 824 C. beijerinckii NCIMB 8052 | glcG, cbei2385, xylR, bdhA, bdhB, ptb, ack, adc | [ |
| 同源重组 | 可精确进行基因编辑 | 同源重组效率很低 | C. acetobutylicum NCIMB 8052 | gutD, spo0A | [ |
| 反筛标记介导的等位基因替换 | 相比纯粹的等位基因替换,效率有所提升 | 受制于第一次单交换效率 | C. acetobutylicum ATCC 824 | adh | [ |
| Ⅰ-SceⅠ归巢内切酶介导的等位基因替换 | 适用范围广,在革兰氏阳性和阴性菌中均可使用 | 设计复杂,操作烦琐,周期长 | C. acetobutylicum ATCC 824 C. beijerinckii NCIMB 8052 | adc, glcG, xylR | [ |
| 噬菌体丝氨酸重组酶介导的位点特异性基因编辑技术 | 适用于大片段DNA快速整合到宿主染色体上 | 受限于附着位点attb/p,应用范围小 | C. ljungdahlii DSM 13528 | thl, crt, bcd, etfB, etfA, hbd, ptb, buk | [ |
| Red/ET重组酶介导的同源重组 | 所需同源臂较短、不受限制性酶切位点限制 | 尚不完善 | C. acetobutylicum SMB009 | ermC | [ |
| CRISPR/Cas9系统介导的基因编辑 | 提高同源重组效率,可实现精确编辑 | 毒性大,很难获得转化子 | C. acetobutylicum DSM792 C. saccharoperbutylacetonicum N1-4 C. cellulovorans C. autoethanogenum DSM10061 | hprK, cac1502, pta, buk, clocel2243, adhE1, ctf,pyrE, 2,3-bdh | [ |
| CRISPR/Cas9n系统介导的基因编辑 | 相比于CRISPR/Cas9毒性有所降低,转化子数目增加 | 仍有毒性,转化子依然偏少 | C. cellulolyticum H10 C. acetobutylicum ATCC 824 C. beijerinckii NCIMB 8052 | pyrF, spo0A | [ |
| CRISPR/dCas9系统介导的基因表达下调 | 相比于CRISPR/Cas9毒性大大降低,易于得到转化子 | 依赖于sgRNA和基因;仅在转录水平调控 | C. pasteurianum ATCC 6013 C. acetobutylicum DSM792 | hprK, glpX | [ |
| 1 | 刘朝全, 姜学峰, 戴家权, 等. 疫情促变局 转型谋发展——2020年国内外油气行业发展概述及2021年展望[J]. 国际石油经济, 2021, 29(1): 28-37. |
| LIU C Q, JIANG X F, DAI J Q, et al. Overview of the domestic and foreign oil and gas industry development in 2020 and outlook for 2021[J]. International Petroleum Economics, 2021, 29(1): 28-37. | |
| 2 | 史硕博, 孟琼宇, 乔玮博, 等. 塑造低碳经济的第三代固碳生物炼制[J]. 合成生物学, 2020, 1(1): 44-59. |
| SHI S B, MENG Q Y, QIAO W B, et al. Establishing carbon dioxide-based third-generation biorefinery for a sustainable low-carbon economy[J]. Synthetic Biology Journal, 2020, 1(1): 44-59. | |
| 3 | PAPOUTSAKIS E T. Engineering solventogenic clostridia[J]. Current Opinion in Biotechnology, 2008, 19(5): 420-429. |
| 4 | PYNE M E, BRUDER M, MOO-YOUNG M, et al. Technical guide for genetic advancement of underdeveloped and intractable Clostridium [J]. Biotechnology Advances, 2014, 32(3): 623-641. |
| 5 | JONES D T, WOODS D R. Acetone-butanol fermentation revisited[J]. Microbiological Reviews, 1986, 50(4): 484-524. |
| 6 | TRACY B P, JONES S W, FAST A G, et al. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications[J]. Current Opinion in Biotechnology, 2012, 23(3): 364-381. |
| 7 | VAN MELLAERT L, BARBÉ S, ANNÉ J. Clostridium spores as anti-tumour agents[J]. Trends in Microbiology, 2006, 14(4): 190-196. |
| 8 | ARORA R, BEHERA S, SHARMA N K, et al. Evaluating the pathway for co-fermentation of glucose and xylose for enhanced bioethanol production using flux balance analysis[J]. Biotechnology and Bioprocess Engineering, 2019, 24(6): 924-933. |
| 9 | BANERJEE S, MISHRA G, ROY A. Metabolic engineering of bacteria for renewable bioethanol production from cellulosic biomass[J]. Biotechnology and Bioprocess Engineering, 2019, 24(5): 713-733. |
| 10 | CHOI Y Y, HONG M E, CHANG W S, et al. Autotrophic biodiesel production from the thermotolerant microalga Chlorella sorokiniana by enhancing the carbon availability with temperature adjustment[J]. Biotechnology & Bioprocess Engineering, 2019, 24(1): 223-231. |
| 11 | KWON S W, PAARI K A, MALAVIYA A, et al. Synthetic biology tools for genome and transcriptome engineering of solventogenic Clostridium [J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 282. |
| 12 | ALEXANDRO E T, ZHANG F. Genome editing using Cas9 nickases[M]. Methods in Enzymology. Volume 546. Elsevier, 2014: 161-174. |
| 13 | 刘家宇, 杨智晗, 杨蕾, 等. 合成生物技术驱动酪丁酸梭菌细胞工厂开发的研究进展[J].合成生物学, 2022, 3(6): 1174. |
| LIU J Y, YANG Z H, YANG L, et al. Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnologies[J]. Synthetic Biology Journal, 2022, 3(6): 1174. | |
| 14 | PYNE M E, MOO-YOUNG M, CHUNG D A, et al. Development of an electrotransformation protocol for genetic manipulation of Clostridium pasteurianum [J]. Biotechnology for Biofuels, 2013, 6(1): 50. |
| 15 | MERMELSTEIN L D, WELKER N E, BENNETT G N, et al. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824[J]. Nature Biotechnology, 1992, 10(2): 190-195. |
| 16 | JENNERT K C B, TARDIF C, YOUNG D I, et al. Gene transfer to Clostridium cellulolyticum ATCC 35319[J]. Microbiology, 2000, 146 (12): 3071-3080. |
| 17 | PURDY D, O'KEEFFE T A T, ELMORE M, et al. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier[J]. Molecular Microbiology, 2002, 46(2): 439-452. |
| 18 | RICHARDS D F, LINNETT P E, OULTRAM J D, et al. Restriction endonucleases in Clostridium pasteurianum ATCC 6013 and Clostridium thermohydrosulfuricum DSM 568[J]. Journal of general microbiology, 1988, 134(12): 3151-3157. |
| 19 | KLAPATCH T R, DEMAIN A L, LYND L R. Restriction endonuclease activity in Clostridium thermocellum and Clostridium thermosaccharolyticum [J]. Applied Microbiology & Biotechnology, 1996, 45(1/2): 127-131. |
| 20 | XUE C, CHENG C. Butanol production by Clostridium [M].Advances in Bioenergy. Volume 4. Amsterdam: Elsevier, 2019: 35-77. |
| 21 | MERMELSTEIN L D, PAPOUTSAKIS E T. In vivo methylation in Escherichia coli by the Bacillus subtilis phage-phi-3T-I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824[J]. Applied & Environmental Microbiology, 1993, 59(4): 1077-1081. |
| 22 | MOLITOR B, KIRCHNER K, HENRICH A W, et al. Expanding the molecular toolkit for the homoacetogen Clostridium ljungdahlii [J]. Scientific Reports, 2016, 6: 31518. |
| 23 | DONG H J, ZHANG Y P, DAI Z J, et al. Engineering Clostridium strain to accept unmethylated DNA[J]. PLoS One, 2010, 5(2): e9038. |
| 24 | GONZÁLEZ-PAJUELO M, MEYNIAL-SALLES I, MENDES F, et al. Microbial conversion of glycerol to 1,3-propanediol: physiological comparison of a natural producer, Clostridium butyricum VPI 3266, and an engineered strain, Clostridium acetobutylicum DG1(pSPD5)[J]. Applied and Environmental Microbiology, 2006, 72(1): 96-101. |
| 25 | HEAP J T, PENNINGTON O J, CARTMAN S T, et al. The ClosTron: a universal gene knock-out system for the genus Clostridium [J]. Journal of Microbiological Methods, 2007, 70(3): 452-464. |
| 26 | LÜTKE-EVERSLOH T. Application of new metabolic engineering tools for Clostridium acetobutylicum [J]. Applied Microbiology and Biotechnology, 2014, 98(13): 5823-5837. |
| 27 | SILLERS R, CHOW A, TRACY B, et al. Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance[J]. Metabolic Engineering, 2008, 10(6): 321-332. |
| 28 | ALSAKER K V, SPITZER T R, PAPOUTSAKIS E T. Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress[J]. Journal of Bacteriology, 2004, 186(7): 1959-1971. |
| 29 | GIRBAL L, VON ABENDROTH G, WINKLER M, et al. Homologous and heterologous overexpression in Clostridium acetobutylicum and characterization of purified clostridial and algal Fe-only hydrogenases with high specific activities[J]. Applied and Environmental Microbiology, 2005, 71(5): 2777-2781. |
| 30 | TOMAS C A, WELKER N E, PAPOUTSAKIS E T. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program[J]. Applied and Environmental Microbiology, 2003, 69(8): 4951-4965. |
| 31 | YU M R, ZHANG Y L, TANG I C, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production[J]. Metabolic Engineering, 2011, 13(4): 373-382. |
| 32 | KOVÁCS K, WILLSON B J, SCHWARZ K, et al. Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824[J]. Biotechnology for Biofuels, 2013, 6(1): 117. |
| 33 | YU M R, DU Y M, JIANG W Y, et al. Effects of different replicons in conjugative plasmids on transformation efficiency, plasmid stability, gene expression and n-butanol biosynthesis in Clostridium tyrobutyricum [J]. Applied Microbiology and Biotechnology, 2012, 93(2): 881-889. |
| 34 | MANI N, DUPUY B. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(10): 5844-5849. |
| 35 | MANI N, LYRAS D, BARROSO L, et al. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression[J]. Journal of Bacteriology, 2002, 184(21): 5971-5978. |
| 36 | LEE J Y, JANG Y S, LEE J, et al. Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production[J]. Biotechnology Journal, 2009, 4(10): 1432-1440. |
| 37 | MATAMOUROS S, ENGLAND P, DUPUY B. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC [J]. Molecular Microbiology, 2007, 64(5): 1274-1288. |
| 38 | GUEDON E, DESVAUX M, PETITDEMANGE H. Improvement of cellulolytic properties of Clostridium cellulolyticum by metabolic engineering[J]. Applied and Environmental Microbiology, 2002, 68(1): 53-58. |
| 39 | DESAI R P, PAPOUTSAKIS E T. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum [J]. Applied and Environmental Microbiology, 1999, 65(3): 936-945. |
| 40 | TUMMALA S B, WELKER N E, PAPOUTSAKIS E T. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum [J]. Journal of Bacteriology, 2003, 185(6): 1923-1934. |
| 41 | SILLERS R, AL-HINAI M A, PAPOUTSAKIS E T. Aldehyde-alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicumfermentations[J]. Biotechnology and Bioengineering, 2009, 102(1): 38-49. |
| 42 | VIDAL J E, CHEN J M, LI J H, et al. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13[J]. PLoS One, 2009, 4(7): e6232. |
| 43 | MULLANY P, WILKS M, TABAQCHALI S. Transfer of Tn916 and Tn916 ΔE into Clostridium difficile: demonstration of a hot-spot for these elements in the C. difficile genome[J]. FEMS Microbiology Letters, 1991, 63(2/3): 191-194. |
| 44 | XIAO H, GU Y, NING Y Y, et al. Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose[J]. Applied and Environmental Microbiology, 2011, 77(22): 7886-7895. |
| 45 | XIAO H, LI Z L, JIANG Y, et al. Metabolic engineering of D-xylose pathway in Clostridium beijerinckii to optimize solvent production from xylose mother liquid[J]. Metabolic Engineering, 2012, 14(5): 569-578. |
| 46 | GU Y, DING Y, REN C, et al. Reconstruction of xylose utilization pathway and regulons in firmicutes[J]. BMC Genomics, 2010, 11: 255. |
| 47 | COOKSLEY C M, ZHANG Y, WANG H Z, et al. Targeted mutagenesis of the Clostridium acetobutylicum acetone-butanol-ethanol fermentation pathway[J]. Metabolic Engineering, 2012, 14(6): 630-641. |
| 48 | WILKINSON S R, YOUNG M. Targeted integration of genes into the Clostridium acetobutylicum chromosome[J]. Microbiology, 1994, 140(1): 89-95. |
| 49 | HEAP J T, EHSAAN M, COOKSLEY C M, et al. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker[J]. Nucleic Acids Research, 2012, 40(8): e59. |
| 50 | ZHANG N, SHAO L J, JIANG Y, et al. I-SceI-mediated scarless gene modification via allelic exchange in Clostridium [J]. Journal of Microbiological Methods, 2015, 108: 49-60. |
| 51 | HUANG H, CHAI C S, YANG S, et al. Phage serine integrase-mediated genome engineering for efficient expression of chemical biosynthetic pathway in gas-fermenting Clostridium ljungdahlii [J]. Metabolic Engineering, 2019, 52: 293-302. |
| 52 | DONG H J, TAO W W, GONG F Y, et al. A functional recT gene for recombineering of Clostridium [J]. Journal of Biotechnology, 2014, 173: 65-67. |
| 53 | BRUDER M R, PYNE M E, MOO-YOUNG M, et al. Extending CRISPR-Cas9 technology from genome editing to transcriptional engineering in the genus Clostridium [J]. Applied and Environmental Microbiology, 2016, 82(20): 6109-6119. |
| 54 | WANG S H, DONG S, WANG P X, et al. Genome editing in Clostridium saccharoperbutylacetonicum N1-4 with the CRISPR-Cas9 system[J]. Applied & Environmental Microbiology, 2017, 83(10): e00233-e00217. |
| 55 | WEN Z Q, MINTON N P, ZHANG Y, et al. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium[J]. Metabolic Engineering, 2017, 39: 38-48. |
| 56 | HUANG H, CHAI C S, LI N, et al. CRISPR/Cas9-based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium[J]. ACS Synthetic Biology, 2016, 5(12): 1355-1361. |
| 57 | NAGARAJU S, DAVIES N K, WALKER D J F, et al. Genome editing of Clostridium autoethanogenum using CRISPR/Cas9[J]. Biotechnology for Biofuels, 2016, 9: 219. |
| 58 | LI Q, CHEN J, MINTON N P, et al. CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii [J]. Biotechnology Journal, 2016, 11(7): 961-972. |
| 59 | XU T, LI Y C, SHI Z, et al. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase[J]. Applied and Environmental Microbiology, 2015, 81(13): 4423-4431. |
| 60 | DORNENBURG J E, DEVITA A M, PALUMBO M J, et al. Widespread antisense transcription in Escherichia coli [J]. mBio, 2010, 1(1): e00024-e00010. |
| 61 | 马欣, 高学文. 转座子随机突变芽孢杆菌的研究进展[J]. 中国生物防治学报, 2015, 31(3): 394-403. |
| MA X, GAO X W. Research progress of random mutagenesis of bacillus by use of transposon[J]. Chinese Journal of Biological Control, 2015, 31(3): 394-403. | |
| 62 | 顾阳, 杨晟, 姜卫红. 产溶剂梭菌分子遗传操作技术研究进展[J]. 生物工程学报, 2013, 29(8): 1133-1145. |
| GU Y, YANG S, JIANG W H. Development in molecular genetic manipulation of solventogenic clostridia[J]. Chinese Journal of Biotechnology, 2013, 29(8): 1133-1145. | |
| 63 | LIU H L, BOUILLAUT L, SONENSHEIN A L, et al. Use of a mariner-based transposon mutagenesis system to isolate Clostridium perfringens mutants deficient in gliding motility[J]. Journal of Bacteriology, 2013, 195(3): 629-636. |
| 64 | LANCKRIET A, TIMBERMONT L, HAPPONEN L J, et al. Generation of single-copy transposon insertions in Clostridium perfringens by electroporation of phage mu DNA transposition complexes[J]. Applied and Environmental Microbiology, 2009, 75(9): 2638-2642. |
| 65 | CARTMAN S T, MINTON N P. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile [J]. Applied and Environmental Microbiology, 2010, 76(4): 1103-1109. |
| 66 | WANG H, ROBERTS A P, LYRAS D, et al. Characterization of the ends and target sites of the novel conjugative transposon Tn5397 from Clostridium difficile: excision and circularization is mediated by the large resolvase, TndX[J]. Journal of Bacteriology, 2000, 182(13): 3775-3783. |
| 67 | ZHANG Y, XU S, CHAI C S, et al. Development of an inducible transposon system for efficient random mutagenesis in Clostridium acetobutylicum [J]. FEMS Microbiology Letters, 2016, 363(8): fnw065. |
| 68 | KUEHNE S A, MINTON N P. ClosTron-mediated engineering of Clostridium [J]. Bioengineered, 2012, 3(4): 247-254. |
| 69 | SHAO L J, HU S Y, YANG Y, et al. Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum [J]. Cell Research, 2007, 17(11): 963-965. |
| 70 | 闻志强, 孙小曼, 汪庆卓, 等. 梭菌正丁醇代谢工程研究进展[J]. 合成生物学, 2021, 2(2): 194-221. |
| WEN Z Q, SUN X M, WANG Q Z, et al. Recent advances in metabolic engineering of clostridia for n-butanol production[J]. Synthetic Biology Journal, 2021, 2(2): 194-221. | |
| 71 | HÖNICKE D, LÜTKE-EVERSLOH T, LIU Z Y, et al. Chemostat cultivation and transcriptional analyses of Clostridium acetobutylicum mutants with defects in the acid and acetone biosynthetic pathways[J]. Applied Microbiology and Biotechnology, 2014, 98(23): 9777-9794. |
| 72 | LIU J, GUO T, WANG D, et al. Enhanced butanol production by increasing NADH and ATP levels in Clostridium beijerinckii NCIMB 8052 by insertional inactivation of Cbei_4110 [J]. Applied Microbiology and Biotechnology, 2016, 100(11): 4985-4996. |
| 73 | JANG Y S, IM J A, CHOI S Y, et al. Metabolic engineering of Clostridium acetobutylicum for butyric acid production with high butyric acid selectivity[J]. Metabolic Engineering, 2014, 23: 165-174. |
| 74 | WEN Z Q, LU M R, LEDESMA-AMARO R, et al. TargeTron technology applicable in solventogenic clostridia: revisiting 12 years' advances[J]. Biotechnology Journal, 2020, 15(1): 1900284. |
| 75 | LAMBOWITZ A M, ZIMMERLY S. Group II introns: mobile ribozymes that invade DNA[J]. Cold Spring Harbor Perspectives in Biology, 2011, 3(8): a003616. |
| 76 | HEAP J T, KUEHNE S A, EHSAAN M, et al. The ClosTron: mutagenesis in Clostridium refined and streamlined[J]. Journal of Microbiological Methods, 2010, 80(1): 49-55. |
| 77 | ARGYROS D A, TRIPATHI S A, BARRETT T F, et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes[J]. Applied and Environmental Microbiology, 2011, 77(23): 8288-8294. |
| 78 | KOHJI K, NORIKO K T, HIROSHI Y, et al. Involvement of RecE exonuclease and RecT annealing protein in DNA double-strand break repair by homologous recombination[J]. Gene, 1994, 138(1/2): 17-25. |
| 79 | HORVATH P, BARRANGOU R. CRISPR/Cas, the immune system of bacteria and archaea[J]. Science, 2010, 327(5962): 167-170. |
| 80 | JANSEN R, VAN EMBDEN J D A, GAASTRA W, et al. Identification of genes that are associated with DNA repeats in prokaryotes[J]. Molecular Microbiology, 2002, 43(6): 1565-1575. |
| 81 | MALI P, YANG L H, ESVELT K M, et al. RNA-guided human genome engineering via Cas9[J]. Science, 2013, 339(6121): 823-826. |
| 82 | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| 83 | WANG Y, ZHANG Z T, SEO S O, et al. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system[J]. Journal of Biotechnology, 2015, 200: 1-5. |
| 84 | WANG Y, ZHANG Z T, SEO S O, et al. Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable "clean" mutant selection in Clostridium beijerinckii as an example[J]. ACS Synthetic Biology, 2016, 5(7): 721-732. |
| 85 | BERNHEIM A, CALVO-VILLAMAÑÁN A, BASIER C, et al. Inhibition of NHEJ repair by type II-A CRISPR-Cas systems in bacteria[J]. Nature Communications, 2017, 8: 2094. |
| 86 | CUI L, BIKARD D. Consequences of Cas9 cleavage in the chromosome of Escherichia coli [J]. Nucleic Acids Research, 2016, 44(9): 4243-4251. |
| 87 | SHUMAN S, GLICKMAN M S. Bacterial DNA repair by non-homologous end joining[J]. Nature Reviews Microbiology, 2007, 5(11): 852-861. |
| 88 | XU T, LI Y C, HE Z L, et al. Cas9 nickase-assisted RNA repression enables stable and efficient manipulation of essential metabolic genes in Clostridium cellulolyticum [J]. Frontiers in Microbiology, 2017, 8: 1744. |
| 89 | STRECKER J, LADHA A, GARDNER Z, et al. RNA-guided DNA insertion with CRISPR-associated transposases[J]. Science, 2019, 365(6448): 48-53. |
| 90 | PYNE M E, BRUDER M R, MOO-YOUNG M, et al. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium [J]. Scientific Reports, 2016, 6: 25666. |
| 91 | ZHANG J, ZONG W M, HONG W, et al. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production[J]. Metabolic Engineering, 2018, 47: 49-59. |
| 92 | ZHOU X Q, WANG X L, LUO H Y, et al. Exploiting heterologous and endogenous CRISPR-Cas systems for genome editing in the probiotic Clostridium butyricum [J]. Biotechnology and Bioengineering, 2021, 118(7): 2448-2459. |
| 93 | WENTZ T G, TREMBLAY B J M, BRADSHAW M, et al. Endogenous CRISPR-Cas systems in group I Clostridium botulinum and Clostridium sporogenes do not directly target the botulinum neurotoxin gene cluster[J]. Frontiers in Microbiology, 2021, 12: 787726. |
| 94 | WALKER J E, LANAHAN A A, ZHENG T Y, et al. Development of both type I-B and type II CRISPR/Cas genome editing systems in the cellulolytic bacterium Clostridium thermocellum [J]. Metabolic Engineering Communications, 2019, 10: eoo116. |
| 95 | HONG W, ZHANG J, CUI G Z, et al. Multiplexed CRISPR-Cpf1-mediated genome editing in Clostridium difficile toward the understanding of pathogenesis of C. difficile infection[J]. ACS Synthetic Biology, 2018, 7(6): 1588-1600. |
| 96 | MERRICK C A, ZHAO J, ROSSER S J. Serine integrases: advancing synthetic biology[J]. ACS Synthetic Biology, 2018, 7(2): 299-310. |
| 97 | BAI H, SUN M X, GHOSH P, et al. Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(18): 7419-7424. |
| 98 | 田雨顺, 罗鹏, 刘秋婷, 等. 细菌重组系统及其应用研究进展[J]. 微生物学通报, 2017, 44(2): 473-482. |
| TIAN Y S, LUO P, LIU Q T, et al. Progress on bacterial recombination systems and their application[J]. Microbiology China, 2017, 44(2): 473-482. | |
| 99 | IYER L M, KOONIN E V, ARAVIND L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redβ, ERF and RAD52[J]. BMC Genomics, 2002, 3: 8. |
| 100 | 王雪. 伯克氏菌重组系统的研究与应用[D]. 济南:山东大学, 2018. |
| WANG X. The research and application of recombineering system in Burkholderiales species[D]. Jinan: Shandong University, 2018. | |
| 101 | SHARAN S K, THOMASON L C, KUZNETSOV S G, et al. Recombineering: a homologous recombination-based method of genetic engineering[J]. Nature Protocols, 2009, 4(2): 206-223. |
| 102 | REISCH C R, PRATHER K L J. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli [J]. Scientific Reports, 2015, 5: 15096. |
| 103 | WANG H H, ISAACS F J, CARR P A, et al. Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894-898. |
| 104 | WARNER J R, REEDER P J, KARIMPOUR-FARD A, et al. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides[J]. Nature Biotechnology, 2010, 28(8): 856-862. |
| 105 | VAN KESSEL J C, HATFULL G F. Recombineering in Mycobacterium tuberculosis [J]. Nature Methods, 2007, 4(2): 147-152. |
| 106 | VAN KESSEL J C, HATFULL G F. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets[J]. Molecular Microbiology, 2008, 67(5): 1094-1107. |
| 107 | VAN PIJKEREN J P, BRITTON R A. High efficiency recombineering in lactic acid bacteria[J]. Nucleic Acids Research, 2012, 40(10): e76. |
| [1] | 董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117. |
| [2] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [3] | 陈盈盈, 刘扬, 史俊杰, 马俊英, 鞠建华. CRISPR/Cas基因编辑及其新兴技术在丝状真菌研究中的系统应用[J]. 合成生物学, 2024, 5(3): 672-693. |
| [4] | 惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671. |
| [5] | 杜瑶, 高宏丹, 刘家坤, 刘孝荣, 邢志浩, 张涛, 马东礼. CRISPR-Cas系统在病原核酸检测中的研究进展[J]. 合成生物学, 2024, 5(1): 202-216. |
| [6] | 许志锰, 谢震. 引导编辑研究进展及其应用[J]. 合成生物学, 2024, 5(1): 1-15. |
| [7] | 陈雅如, 曹英秀, 宋浩. 电活性微生物基因编辑与转录调控技术进展与应用[J]. 合成生物学, 2023, 4(6): 1281-1299. |
| [8] | 王甜甜, 朱虹, 杨琛. 蓝细菌CRISPRa系统的开发及其代谢工程应用[J]. 合成生物学, 2023, 4(4): 824-839. |
| [9] | 马孟丹, 尚梦宇, 刘宇辰. CRISPR-Cas9系统在肿瘤生物学中的应用及前景[J]. 合成生物学, 2023, 4(4): 703-719. |
| [10] | 林继聪, 邹根, 刘宏民, 魏勇军. CRISPR/Cas基因组编辑技术在丝状真菌次级代谢产物合成中的应用[J]. 合成生物学, 2023, 4(4): 738-755. |
| [11] | 马孟丹, 刘宇辰. 合成生物学在疾病信息记录与实时监测中的应用潜力[J]. 合成生物学, 2023, 4(2): 301-317. |
| [12] | 柳柯, 林桂虹, 刘坤, 周伟, 王风清, 魏东芝. CRISPR/Cas系统的挖掘、改造与功能拓展[J]. 合成生物学, 2023, 4(1): 47-66. |
| [13] | 潘颖佳, 夏思杨, 董昌, 蔡谨, 连佳长. 基因增变器驱动的酿酒酵母基因组连续进化[J]. 合成生物学, 2023, 4(1): 225-240. |
| [14] | 滕小龙, 史硕博. CRISPR/Cas9系统在基因组编辑中的优化与发展[J]. 合成生物学, 2023, 4(1): 67-85. |
| [15] | 梁丽亚, 刘嵘明. 靶向DNA的Ⅱ类CRISPR/Cas系统的蛋白工程化改造[J]. 合成生物学, 2023, 4(1): 86-101. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||