Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (6): 1017-1029.DOI: 10.12211/2096-8280.2021-011
• Invited Review • Previous Articles Next Articles
Progress in construction and applications of methanotrophic cell factory for chemicals biosynthesis
GUO Shuqi1, JIAO Ziyue1, FEI Qiang1,2
- 1.School of Chemical Engineering and Technology,Xi’an Jiaotong University,Xi’an 710049,Shaanxi,China
2.Shaanxi Key Laboratory of Energy Chemical Process Intensification,Xi’an Jiaotong University,Xi’an 710049,Shaanxi,China
-
Received:2021-01-25Revised:2021-04-30Online:2022-01-21Published:2021-12-31 -
Contact:FEI Qiang
基于化学品生物合成的嗜甲烷菌人工细胞构建及应用进展
郭树奇1, 焦子悦1, 费强1,2
- 1.西安交通大学化学工程与技术学院,陕西 西安 710049
2.陕西省能源化工过程强化重点实验室,陕西 西安 710049
-
通讯作者:费强 -
作者简介:郭树奇 (1989—),男,博士、助理教授。研究方向为微生物代谢工程及合成生物学。E-mail:shuqguo@xjtu.edu.cn费强 (1980—),男,教授,博士生导师。研究方向为围绕构建人工细胞将一碳气体高效转化为平台化学品和生物能源,并对其放大工艺进行技术经济可行性分析。E-mail:feiqiang@xjtu.edu.cn -
基金资助:国家重点研发计划(2018YFA0901500);国家自然科学基金(21878241);陕西省重点研发计划(2021SF-103)
CLC Number:
Cite this article
GUO Shuqi, JIAO Ziyue, FEI Qiang. Progress in construction and applications of methanotrophic cell factory for chemicals biosynthesis[J]. Synthetic Biology Journal, 2021, 2(6): 1017-1029.
郭树奇, 焦子悦, 费强. 基于化学品生物合成的嗜甲烷菌人工细胞构建及应用进展[J]. 合成生物学, 2021, 2(6): 1017-1029.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-011
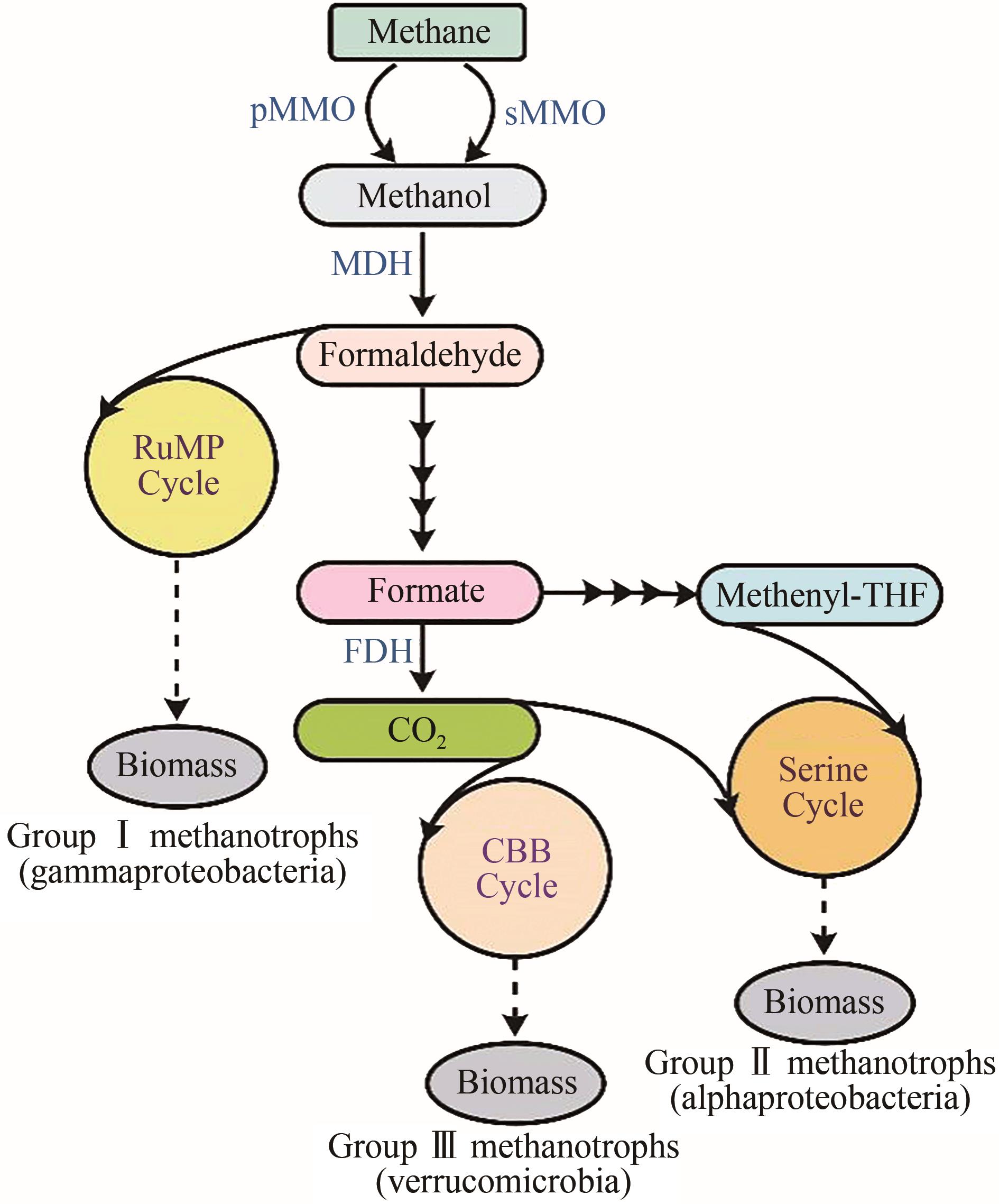
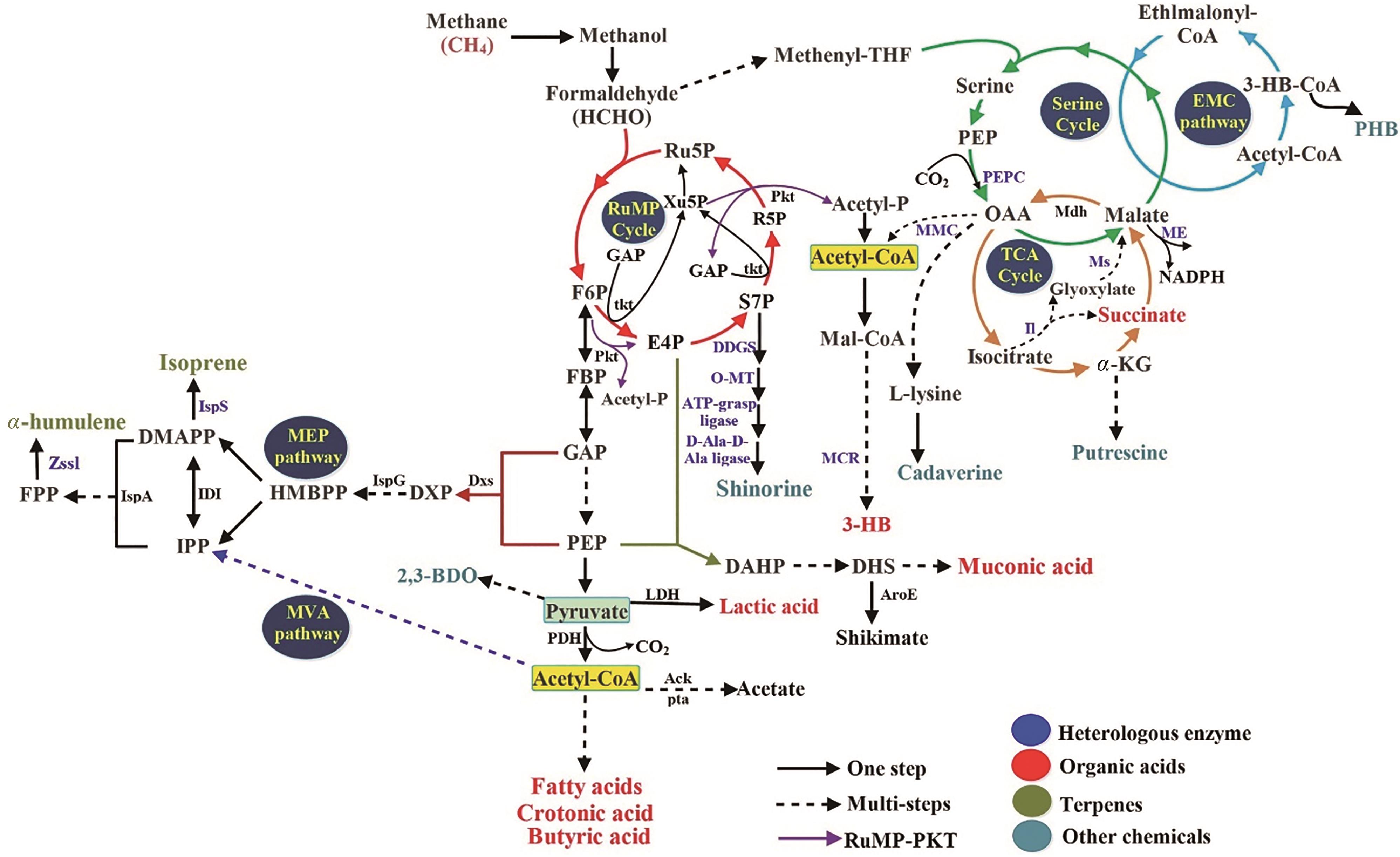
Fig. 1 The pathways of methane metabolism in methanotrophs[8, 27] pMMO—particulate methane monooxygenase; sMMO—soluble methane monooxygenase; MDH—methanol dehydrogenase; FDH—formate dehydrogenase
| 功能 | 质粒 | 筛选标记 | 菌株 | 参考文献 |
|---|---|---|---|---|
| 基因复制 | pBHR1 | Km, Cm | Methylomonas sp. 16a | [ |
| pAWP78 | Km | M. buryatense 5G | [ | |
| 基因表达 | pAWP89 | Km | M. buryatense 5G | [ |
| pCAH01:emGFP | Km | M. buryatense 5G | [ | |
| 基因敲除 | pK18mobsacB | Km, SacB | Methylococcus capsulatus (Bath) | [ |
| pCM184 | Gm, SacB | Methylomicrobium alcaliphilum 20Z | [ | |
| 基因转移技术 | ||||
| 接合转移 | pRK2013 | Km | Methylomonas sp. 16a | [ |
| pBHR1 | Km | Methylomonas sp. 16a | [ | |
| 电转化 | pAWP89 (线性化) | Km | M. buryatense 5G | [ |
| pheSAG (线性化DNA片段) | Km | M. buryatense 5G | [ |
Tab. 1 The genetic tool used for metabolic engineering of methanotrophs
| 功能 | 质粒 | 筛选标记 | 菌株 | 参考文献 |
|---|---|---|---|---|
| 基因复制 | pBHR1 | Km, Cm | Methylomonas sp. 16a | [ |
| pAWP78 | Km | M. buryatense 5G | [ | |
| 基因表达 | pAWP89 | Km | M. buryatense 5G | [ |
| pCAH01:emGFP | Km | M. buryatense 5G | [ | |
| 基因敲除 | pK18mobsacB | Km, SacB | Methylococcus capsulatus (Bath) | [ |
| pCM184 | Gm, SacB | Methylomicrobium alcaliphilum 20Z | [ | |
| 基因转移技术 | ||||
| 接合转移 | pRK2013 | Km | Methylomonas sp. 16a | [ |
| pBHR1 | Km | Methylomonas sp. 16a | [ | |
| 电转化 | pAWP89 (线性化) | Km | M. buryatense 5G | [ |
| pheSAG (线性化DNA片段) | Km | M. buryatense 5G | [ |
| 菌株 | 产物种类 | 代谢途径/前体 | 产物 | 产量 | 参考文献 |
|---|---|---|---|---|---|
| M. buryatense 5GB1C[ | 有机酸 | 丙酮酸 | L-乳酸 | 800 mg/L | [ |
| 有机酸 | 丙酮酸 | L-乳酸 | 600 mg/L | [ | |
| 有机酸 | 乙酰辅酶A | 丁烯酸 | 70 mg/L | [ | |
| 有机酸 | 乙酰辅酶A | 丁酸 | 40 mg/L | [ | |
| 有机酸 | 莽草酸途径 | 黏糠酸 | 12.4 mg/L | [ | |
| 有机酸 | 乙酰辅酶A | 脂肪酸 | 111 mg/g DCW | [ | |
| M. alcaliphilum 20Z[ | 生物醇 | 丙酮酸 | 2,3-丁二醇 | 86.2 mg/L | [ |
| 生物醇 | 丙酮酸 | 2,3-丁二醇 | 361.3 mg/L | [ | |
| 有机酸 | 乙酰辅酶A | 3-羟基丙酸 | 196.3 mg/L | [ | |
| 有机酸 | 莽草酸途径 | 黏糠酸 | 0.75 mg/L | [ | |
| 萜类 | MEP 途径 | α-蛇麻烯 | 0.75 mg/g DCW | [ | |
| — | TCA循环 | 腐胺 | 98.08 mg/L | [ | |
| 有机酸 | 丙酮酸 | 乳酸 | 0.027 g/(g DCW·h) | [ | |
| — | RuMP 循环 | Shinorine | 31 mg/L | [ | |
| M. capsulatus Bath[ | 有机酸 | 莽草酸途径 | 黏糠酸 | 1.0 mg/L | [ |
| 萜类 | MEP 途径 | 异戊二烯 | 10 mg/L | [ | |
| 生物醇 | TCA 循环 | 1,4-丁二醇 | — | [ | |
| 生物醇 | 丙酮酸 | 异丙醇 | 220 mg/L① | [ | |
| 生物醇 | 丙酮酸 | 异丙醇 | 1 mg/L | [ | |
| Methylomonas sp. DH-1[ | 有机酸 | TCA 循环 | 琥珀酸 | 195 mg/L | [ |
| 有机酸 | 丙酮酸 | D-乳酸 | 1190 mg/L | [ | |
| Methylosinus trichosporium OB3b[ | 有机酸 | 乙酰辅酶A | 3-羟基丙酸 | 60.6 mg/L | [ |
| — | TCA 循环 | 尸胺 | 283.6 mg/L | [ | |
| Methylomonas sp. 16a[ | 萜类 | MEP 途径 | 柠檬烯 | 0.5 mg/L | [ |
| 萜类 | MEP 途径 | 法尼烯 | — | [ | |
| 萜类 | MEP 途径 | 虾青素 | 2.4 mg/g DCW | [ | |
| 萜类 | MEP 途径 | 虾青素 | 2.0 mg/g DCW | [ |
Tab. 2 Biosynthesis of various chemicals and biofuels from methane by methanotrophic cell factories
| 菌株 | 产物种类 | 代谢途径/前体 | 产物 | 产量 | 参考文献 |
|---|---|---|---|---|---|
| M. buryatense 5GB1C[ | 有机酸 | 丙酮酸 | L-乳酸 | 800 mg/L | [ |
| 有机酸 | 丙酮酸 | L-乳酸 | 600 mg/L | [ | |
| 有机酸 | 乙酰辅酶A | 丁烯酸 | 70 mg/L | [ | |
| 有机酸 | 乙酰辅酶A | 丁酸 | 40 mg/L | [ | |
| 有机酸 | 莽草酸途径 | 黏糠酸 | 12.4 mg/L | [ | |
| 有机酸 | 乙酰辅酶A | 脂肪酸 | 111 mg/g DCW | [ | |
| M. alcaliphilum 20Z[ | 生物醇 | 丙酮酸 | 2,3-丁二醇 | 86.2 mg/L | [ |
| 生物醇 | 丙酮酸 | 2,3-丁二醇 | 361.3 mg/L | [ | |
| 有机酸 | 乙酰辅酶A | 3-羟基丙酸 | 196.3 mg/L | [ | |
| 有机酸 | 莽草酸途径 | 黏糠酸 | 0.75 mg/L | [ | |
| 萜类 | MEP 途径 | α-蛇麻烯 | 0.75 mg/g DCW | [ | |
| — | TCA循环 | 腐胺 | 98.08 mg/L | [ | |
| 有机酸 | 丙酮酸 | 乳酸 | 0.027 g/(g DCW·h) | [ | |
| — | RuMP 循环 | Shinorine | 31 mg/L | [ | |
| M. capsulatus Bath[ | 有机酸 | 莽草酸途径 | 黏糠酸 | 1.0 mg/L | [ |
| 萜类 | MEP 途径 | 异戊二烯 | 10 mg/L | [ | |
| 生物醇 | TCA 循环 | 1,4-丁二醇 | — | [ | |
| 生物醇 | 丙酮酸 | 异丙醇 | 220 mg/L① | [ | |
| 生物醇 | 丙酮酸 | 异丙醇 | 1 mg/L | [ | |
| Methylomonas sp. DH-1[ | 有机酸 | TCA 循环 | 琥珀酸 | 195 mg/L | [ |
| 有机酸 | 丙酮酸 | D-乳酸 | 1190 mg/L | [ | |
| Methylosinus trichosporium OB3b[ | 有机酸 | 乙酰辅酶A | 3-羟基丙酸 | 60.6 mg/L | [ |
| — | TCA 循环 | 尸胺 | 283.6 mg/L | [ | |
| Methylomonas sp. 16a[ | 萜类 | MEP 途径 | 柠檬烯 | 0.5 mg/L | [ |
| 萜类 | MEP 途径 | 法尼烯 | — | [ | |
| 萜类 | MEP 途径 | 虾青素 | 2.4 mg/g DCW | [ | |
| 萜类 | MEP 途径 | 虾青素 | 2.0 mg/g DCW | [ |
| 1 | FEI Q, PIENKOS P T. Bioconversion of methane for value-added products[M]//SANI R K, RATHINAM N K. Extremophilic microbial processing of lignocellulosic feedstocks to biofuels, value-added products, and usable power. Cham: Springer, 2018: 145-162. |
| 2 | YVON-DUROCHER G, ALLEN A P, BASTVIKEN D, et al. Methane fluxes show consistent temperature dependence across microbial to ecosystem scales[J]. Nature, 2014, 507(7493): 488-491. |
| 3 | EPA. Understanding global warming potentials, 2017[EB/OL]. . |
| 4 | FELDMAN D R, COLLINS W D, BIRAUD S C, et al. Observationally derived rise in methane surface forcing mediated by water vapour trends[J]. Nature Geoscience, 2018, 11(4): 238-243. |
| 5 | BOUSQUET P, CIAIS P, MILLER J B, et al. Contribution of anthropogenic and natural sources to atmospheric methane variability[J]. Nature, 2006, 443(7110): 439-443. |
| 6 | EPA. Inventory of U.S. Greenhouse gas emissions and sinks: 1990-2013, 2015[EB/OL]. . |
| 7 | HWANG I Y, HUR D H, LEE J H, et al. Batch conversion of methane to methanol using Methylosinus trichosporium OB3b as biocatalyst[J]. Journal of Microbiology and Biotechnology, 2015, 25(3): 375-380. |
| 8 | KALYUZHNAYA M G, PURI A W, LIDSTROM M E. Metabolic engineering in methanotrophic bacteria[J]. Metabolic Engineering, 2015, 29: 142-152. |
| 9 | 胡礼珍, 王佳, 袁波, 等. 碳一气体生物利用进展[J]. 生物加工过程, 2017, 15(6): 17-25. |
| HU L Z, WANG J, YUAN B, et al. Production of biofuels and chemicals from C1 gases by microorganisms: status and prospects[J]. Chinese Journal of Bioprocess Engineering, 2017, 15(6): 17-25. | |
| 10 | LIU Y C, HE X R, ZHU P P, et al. pheSAG based rapid and efficient markerless mutagenesis in Methylotuvimicrobium [J]. Frontiers in Microbiology, 2020, 11: 441. |
| 11 | JEON Y C, ANH D N, LEE E Y. Bioproduction of Isoprenoids and other secondary metabolites using methanotrophic bacteria as an alternative microbial cell factory option: current stage and future aspects[J]. Catalysts, 2019, 9(11): 883. |
| 12 | CANTERA S, BORDEL S, LEBRERO R, et al. Bio-conversion of methane into high profit margin compounds: an innovative, environmentally friendly and cost-effective platform for methane abatement[J]. World Journal of Microbiology and Biotechnology, 2019, 35(1): 16. |
| 13 | NGUYEN A D, LEE E Y. Engineered methanotrophy: a sustainable solution for methane-based industrial biomanufacturing[J]. Trends in Biotechnology, 2021, 39 (4): 381-396. |
| 14 | NGUYEN A D, HWANG I Y, LEE O K, et al. Systematic metabolic engineering of Methylomicrobium alcaliphilum 20Z for 2,3-butanediol production from methane[J]. Metabolic Engineering, 2018, 47: 323-333. |
| 15 | NGUYEN D T N, LEE O K, LIM C, et al. Metabolic engineering of type Ⅱ methanotroph, Methylosinus trichosporium OB3b, for production of 3-hydroxypropionic acid from methane via a malonyl-CoA reductase-dependent pathway[J]. Metabolic Engineering, 2020, 59: 142-150. |
| 16 | FU Y F, LI Y, LIDSTROM M. The oxidative TCA cycle operates during methanotrophic growth of the Type I methanotroph Methylomicrobium buryatense 5GB1[J]. Metabolic Engineering, 2017, 42: 43-51. |
| 17 | 陆吉学, 王世珍, 方柏山. 生物分子机器——甲烷单加氧酶的研究进展[J]. 生物工程学报, 2015, 31(7): 1015-1023. |
| LU J X, WANG S Z, FANG B S, et al. Advances in biomolecular machine: methane monooxygenases[J].Chinese Journal of Biotechnology, 2015, 31(7): 1015-1023. | |
| 18 | 韩冰, 苏涛, 李信, 等. 甲烷氧化菌及甲烷单加氧酶的研究进展[J]. 生物工程学报, 2008, 24(9): 1511-1519. |
| HAN B, SU T, LI X, et al. Research progresses of methanotrophs and methane monooxygenases[J]. Chinese Journal of Biotechnology, 2008, 24(9): 1511-1519. | |
| 19 | KO Y S, KIM J W, LEE J A, et al. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production[J]. Chemical Society Reviews, 2020, 49(14): 4615-4636. |
| 20 | LEE O K, NGUYEN D T N, LEE E Y. Metabolic engineering of methanotrophs for the production of chemicals and fuels[M]// LEE E Y. Methanotrophs. Cham: Springer, 2019: 163-203. |
| 21 | CHOI K R, JANG W D, YANG D, et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering[J]. Trends in Biotechnology, 2019, 37(8): 817-837. |
| 22 | ANTONIEWICZ M R. Synthetic methylotrophy: strategies to assimilate methanol for growth and chemicals production[J]. Current Opinion in Biotechnology, 2019, 59: 165-174. |
| 23 | HWANG I Y, NGUYEN A D, NGUYEN T T, et al. Biological conversion of methane to chemicals and fuels: technical challenges and issues[J]. Applied Microbiology and Biotechnology, 2018, 102(7): 3071-3080. |
| 24 | CLOMBURG J M, CRUMBLEY A M, GONZALEZ R. Industrial biomanufacturing: the future of chemical production[J]. Science, 2017, 355: aag08046320. |
| 25 | FEI Q, GUARNIERI M T, TAO L, et al. Bioconversion of natural gas to liquid fuel: opportunities and challenges[J]. Biotechnology Advances, 2014, 32(3): 596-614. |
| 26 | MURRELL J C, JETTEN M S M. The microbial methane cycle[J]. Environmental Microbiology Reports, 2009, 1(5): 279-284. |
| 27 | LEE O K, HUR D H, NGUYEN D T N, et al. Metabolic engineering of methanotrophs and its application to production of chemicals and biofuels from methane[J]. Biofuels, Bioproducts and Biorefining, 2016, 10(6): 848-863. |
| 28 | SEMRAU J D, DISPIRITO A A, YOON S. Methanotrophs and copper[J]. FEMS Microbiology Reviews, 2010, 34: 496-531. |
| 29 | NARIYA S, KALYUZHNAYA M G. Diversity, physiology, and biotechnological potential of halo(alkali)philic methane-consuming bacteria[M]// LEE E Y. Methanotrophs. Cham: Springer, 2019: 139-161. |
| 30 | 郭树奇, 费强. 甲烷生物利用及嗜甲烷菌的工程改造[J]. 生物工程学报, 2021,37(3):816-830. |
| GUO S Q, FEI Q. Bioconversion of methane by metabolically engineered methanotrophs[J]. Chinese Journal of Biotechnology, 2021, 37(3): 816-830. | |
| 31 | GROOM J D, FORD S M, PESESKY M W, et al. A mutagenic screen identifies a tonb-dependent receptor required for the lanthanide metal switch in the type I methanotroph "Methylotuvimicrobium buryatense" 5GB1C[J]. Journal of Bacteriology, 2019, 201(15): e00120-19. |
| 32 | LEE J K, KIM S, KIM W, et al. Efficient production of D-lactate from methane in a lactate-tolerant strain of Methylomonas sp. DH-1 generated by adaptive laboratory evolution[J]. Biotechnology for Biofuels, 2019, 12: 234. |
| 33 | GARG S, CLOMBURG J M, GONZALEZ R. A modular approach for high-flux lactic acid production from methane in an industrial medium using engineered Methylomicrobium buryatense 5GB1[J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(6): 379-391. |
| 34 | PURI A W, OWEN S, CHU F, et al. Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense [J]. Applied and Environmental Microbiology, 2015, 81(5): 1775-1781. |
| 35 | SIRAJUDDIN S, ROSENZWEIG A C. Enzymatic oxidation of methane[J]. Biochemistry, 2015, 54(14): 2283-2294. |
| 36 | SHARPE P L, DICOSIMO D, BOSAK M D, et al. Use of transposon promoter-probe vectors in the metabolic engineering of the obligate methanotroph Methylomonas sp. strain 16a for enhanced C40 carotenoid synthesis[J]. Applied and Environmental Microbiology, 2007, 73(6): 1721-1728. |
| 37 | HENARD C A, SMITH H, DOWE N, et al. Bioconversion of methane to lactate by an obligate methanotrophic bacterium[J]. Scientific Reports, 2016, 6: 21585. |
| 38 | CSÁKI R, BODROSSY L, KLEM J, et al. Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis[J]. Microbiology (Reading), 2003, 149(7): 1785-1795. |
| 39 | NGUYEN A D, CHAU T H T, LEE E Y. Methanotrophic microbial cell factory platform for simultaneous conversion of methane and xylose to value-added chemicals[J]. Chemical Engineering Journal, 2021, 420: 127632. |
| 40 | TAO L, SEDKOVA N, YAO H, et al. Expression of bacterial hemoglobin genes to improve astaxanthin production in a methanotrophic bacterium Methylomonas sp.[J]. Applied Microbiology and Biotechnology, 2007, 74(3): 625-633. |
| 41 | YAN X, CHU F, PURI A W, et al. Electroporation-based genetic manipulation in type I Methanotrophs [J]. Applied and Environmental Microbiology, 2016, 82(7): 2062-2069. |
| 42 | TAPSCOTT T, MICHAEL T G, HENARD C A. Development of a CRISPR/Cas9 system for Methylococcus capsulatus in vivo gene editing.[J]. Applied and Environmental Microbiology, 2019, 85(11): e00340-19. |
| 43 | KHMELENINA V N, BECK D A C, MUNK C, et al. Draft genome sequence of Methylomicrobium buryatense strain 5G, a haloalkaline-tolerant methanotrophic bacterium[J]. Genome Announcements, 2013, 1(4): e00053-13. |
| 44 | GARG S, WU H, CLOMBURG J M, et al. Bioconversion of methane to C-4 carboxylic acids using carbon flux through acetyl-CoA in engineered Methylomicrobium buryatense 5GB1C[J]. Metabolic Engineering, 2018, 48: 175-183. |
| 45 | HENARD C A, AKBERDIN I R, KALYUZHNAYA M G, et al. Muconic acid production from methane using rationally-engineered methanotrophic biocatalysts[J]. Green Chemistry, 2019, 21(24): 6731-6737. |
| 46 | DEMIDENKO A, AKBERDIN I R, ALLEMANN M, et al. Fatty acid biosynthesis pathways in Methylomicrobium buryatense 5G(B1)[J]. Frontiers in Microbiology, 2017, 7: 2167. |
| 47 | VUILLEUMIER S, KHMELENINA V N, BRINGEL F, et al. Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z[J]. Journal of Bacteriology, 2012, 194(2): 551-552. |
| 48 | NGUYEN A D, KIM D, LEE E Y. Unlocking the biosynthesis of sesquiterpenoids from methane via the methylerythritol phosphate pathway in methanotrophic bacteria, using α-humulene as a model compound[J]. Metabolic Engineering, 2020, 61: 69-78. |
| 49 | NGUYEN L T, LEE E Y. Biological conversion of methane to putrescine using genome-scale model-guided metabolic engineering of a methanotrophic bacterium Methylomicrobium alcaliphilum 20Z[J]. Biotechnology for Biofuels, 2019, 12(1): 147. |
| 50 | HENARD C A, FRANKLIN T G, YOUHENNA B, et al. Biogas biocatalysis: methanotrophic bacterial cultivation, metabolite profiling, and bioconversion to lactic acid[J]. Frontiers in Microbiology, 2018, 9: 2610. |
| 51 | WARD N, LARSEN Q, SAKWA J, et al. Genomic Insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath)[J]. PLoS Biology, 2004, 2(10): 1616-1628. |
| 52 | LEONARD E, MINSHULL J, NESS J, et al. Compositions and methods for biological production of isoprene: USWO/2014/138419[P]. 2014-09-12. |
| 53 | HELD M A, ZGAO X, CHAO L Y, et al. New genetically modified microorganism comprising a heterologous gene under the control of a molecular switch, useful for making a multicarbon product, 2,3-butanediol, 1,4-butanediol, and isobutyraldehyde: US 0040344[P]. 2020-02-06. |
| 54 | COLEMAN W J, VIDANES G M, COTTAREL G, et al. Biological conversion of multi-carbon compounds from methane: US20140273128[P]. 2014-09-18. |
| 55 | NGUYEN A D, HWANG I Y, LEE O K, et al. Functional analysis of Methylomonas sp. DH-1 genome as a promising biocatalyst for bioconversion of methane to valuable chemicals[J]. Catalysts, 2018, 8(3): 117. |
| 56 | NGUYEN D T N, LEE O K, HADIYATI S, et al. Metabolic engineering of the type Ⅰ methanotroph Methylomonas sp. DH-1 for production of succinate from methane[J]. Metabolic Engineering, 2019, 54: 170-179. |
| 57 | STEIN L Y, YOON S, SEMRAU J D, et al. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b[J]. Journal of Bacteriology, 2010, 192(24): 6497-6498. |
| 58 | THINGUYEN T, KYUNGLEE O, SANZHARNAIZABEKOV, et al. Bioconversion of methane to cadaverine and lysine using an engineered type Ⅱ methanotroph, Methylosinus trichosporium OB3b[J]. Green Chemistry, 2020, 22, 7803-7811. |
| 59 | DICOSIMO D J, KOFFAS M, ODOM J M, et al. Production of cyclic terpenoids: US6818424[P]. 2004-11-16. |
| 60 | YE R W, YAO H, STEAD K, et al. Construction of the astaxanthin biosynthetic pathway in a methanotrophic bacterium Methylomonas sp strain 16a[J]. Journal of Industrial Microbiology Biotechnology, 2007, 34(4): 289-299. |
| 61 | HENARD C A, SMITH H K, GUARNIERI M T. Phosphoketolase overexpression increases biomass and lipid yield from methane in an obligate methanotrophic biocatalyst[J]. Metabolic Engineering, 2017, 41: 152-158. |
| 62 | BOGORAD I W, LIN T S, LIAO J C. Synthetic non-oxidative glycolysis enables complete carbon conservation[J]. Nature, 2013, 502(7473): 693-697. |
| 63 | CHINEN A, KOZLOV Y I, HARA Y, et al. Innovative metabolic pathway design for efficient l-glutamate production by suppressing CO2 emission[J]. Journal of Bioscience and Bioengineering, 2007, 103(3): 262-269. |
| 64 | HENARD C A, FREED E F, GUARNIERI M T. Phosphoketolase pathway engineering for carbon-efficient biocatalysis[J]. Current Opinion in Biotechnology, 2015, 36: 183-188. |
| 65 | MEADOWS A L, HAWKINS K M, TSEGAYE Y, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production[J]. Nature, 2016, 537(7622): 694-697. |
| 66 | NIU F X, LU Q, BU Y F, et al. Metabolic engineering for the microbial production of isoprenoids: carotenoids and isoprenoid-based biofuels[J]. Synthetic and Systems Biotechnology, 2017, 2(3):167-175. |
| 67 | AKBERDIN I R, THOMPSON M, HAMILTON R, et al. Methane utilization in Methylomicrobium alcaliphilum 20Z(R): a systems approach[J]. Scientific Reports, 2018, 8(1): 2512. |
| 68 | SUN L, GONG M Y, LÜ X, et al. Current advance in biological production of short-chain organic acid[J]. Applied Microbiology and Biotechnology, 2020, 104(21): 9109-9124. |
| 69 | MCANULTY M J, POOSARLA V G, LI J, et al. Metabolic engineering of Methanosarcina acetivorans for lactate production from methane[J]. Biotechnology and Bioengineering, 2017, 114(4): 852-861. |
| 70 | KIM J W, KO Y S, CHAE T U, et al. High-level production of 3-hydroxypropionic acid from glycerol as a sole carbon source using metabolically engineered Escherichia coli [J]. Biotechnology and Bioengineering, 2020, 117(7): 2139-2152. |
| 71 | WANG L, ZONG Z, LIU Y, et al. Metabolic engineering of Yarrowia lipolytica for the biosynthesis of crotonic acid[J]. Bioresource Technology, 2019, 287: 121484. |
| 72 | AHN J H, SEO H, PARK W, et al. Enhanced succinic acid production by mannheimia employing optimal malate dehydrogenase[J]. Nature Communications, 2020, 11(1): 1970. |
| 73 | MENG X, YANG J M, CAO Y J, et al. Increasing fatty acid production in E. coli by simulating the lipid accumulation of oleaginous microorganisms[J]. Journal of Industrial Microbiology Biotechnology, 2011, 38(8): 919-925. |
| 74 | ALBER B E. Biotechnological potential of the ethylmalonyl-CoA pathway[J]. Applied Microbiology and Biotechnology, 2011, 89(1): 17-25. |
| 75 | DAVIS M S, CRONAN J E. Inhibition of Escherichia coli acetyl coenzyme a carboxylase by acyl-acyl carrier protein[J]. Journal of Bacteriology, 2001, 183(4):1499-1503. |
| 76 | KIM S, CHEONG S, GONZALEZ R. Engineering Escherichia coli for the synthesis of short-and medium-chain α,β-unsaturated carboxylic acids[J]. Metabolic Engineering, 2016, 36: 90-98. |
| 77 | CLOMBURG J M, VICK J E, BLANKSCHIEN M D, et al. A synthetic biology approach to engineer a functional reversal of the β-oxidation cycle[J]. ACS Synthetic Biology, 2012, 1(11): 541-554. |
| 78 | AKBERDIN I R, COLLINS D A, HAMILTON R, et al. Rare earth elements alter redox balance in Methylomicrobium alcaliphilum 20Z(R)[J]. Frontiers in Microbiology, 2018, 9: 2735. |
| 79 | BORDEL S R Y H. Genome scale metabolic modeling reveals the metabolic potential of three Type II methanotrophs of the genus Methylocystis [J]. Metabolic Engineering, 2019, 54: 191-199. |
| 80 | FU Y, HE L, REEVE J, et al. Core metabolism shifts during growth on methanol versus methane in the methanotroph Methylomicrobium buryatense 5GB1[J]. mBio, 2019, 10(2): e00406-19. |
| 81 | NGUYEN A D, PARK J Y, HWANG I Y, et al. Genome-scale evaluation of core one-carbon metabolism in gammaproteobacterial methanotrophs grown on methane and methanol[J]. Metabolic Engineering, 2020, 57: 1-12. |
| 82 | HU L Z, YANG Y, YAN X, et al. Molecular mechanism associated with the impact of methane/oxygen gas supply ratios on cell growth of Methylomicrobium buryatense 5GB1 through RNA-Seq[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 263. |
| 83 | HAKOBYAN A, ZHU J, GLATTER T, et al. Hydrogen utilization by Methylocystis sp. strain SC2 expands the known metabolic versatility of type IIa methanotrophs[J]. Metabolic Engineering, 2020, 61:181-196. |
| 84 | FEI Q, PURI A W, SMITH H, et al. Enhanced biological fixation of methane for microbial lipid production by recombinant Methylomicrobium buryatense [J]. Biotechnology for Biofuels, 2018, 11: 129. |
| 85 | DONALDSON G K, HOLLANDS K, PICATAGGIO S K. Biocatalyst for conversion of methane and methanol to isoprene: US20150225743[P]. 2015-08-13. |
| 86 | REN Y Y, LIU S S, JIN G J, et al. Microbial production of limonene and its derivatives: achievements and perspectives.[J]. Biotechnology Advances, 2020, 15(44):107628. |
| 87 | ALEMDAR S, HARTWIG S, FRISTER T, et al. Heterologous expression, purification, and biochemical characterization of α-humulene synthase from Zingiber zerumbet Smith[J]. Applied Biochemistry and Biotechnology, 2016, 178(3): 474-489. |
| 88 | GUO W, LI D, HE R, et al. Synthesizing value-added products from methane by a new Methylomonas [J]. Journal of Applied Microbiology, 2017, 123(5): 1214-1227. |
| 89 | LIANG L Y, LIU R M, FREED E F, et al. Synthetic biology and metabolic engineering employing Escherichia coli for C2-C6 bioalcohol production[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 710. |
| 90 | JANG Y S, KIM B, SHIN J H, et al. Bio-based production of C2-C6 platform chemicals[J]. Biotechnology Bioengineering, 2012, 109(10):2437-2459. |
| 91 | BALSKUS E P, WALSH C T. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria[J]. Science, 2010, 329(5999): 1653-1656. |
| 92 | PARK S H, LEE K, JANG J W, et al. Metabolic engineering of Saccharomyces cerevisiae for production of shinorine, a sunscreen material, from xylose[J]. ACS Synthetic Biology, 2019, 8(2): 346-357. |
| 93 | GONZALEZ J E, LONG C P, ANTONIEWICZ M R. Comprehensive analysis of glucose and xylose metabolism in Escherichia coli under aerobic and anaerobic conditions by 13C metabolic flux analysis[J]. Metabolic Engineering, 2017, 39: 9-18. |
| 94 | GAO J Y, YOU F Q. Design and optimization of shale gas energy systems: overview, research challenges, and future directions[J]. Computers Chemical Engineering, 2017, 106(11): 699-718. |
| 95 | 王红秋, 乔明, 郑轶丹. 美“页岩气化工”重塑全球化工产业链[J]. 中国石油企业, 2017(3): 73-74. |
| WANG H Q, QIAO M, ZHENG Y D. The "shale gas chemical industry" of USA reshapes global chemical industry chain[J]. China Petroleum Enterprise, 2017(3): 73-74. | |
| 96 | 金瑞庭. 当前国际大宗商品价格走势及2020年展望[J]. 中国经贸导刊, 2020(9): 24-25. |
| JIN R T. Current international commodity price trends and prospects for 2020[J]. China Economic Trade Herald, 2020(9): 24-25. | |
| 97 | KALYUZHNAYA M G, YANG S, ROZOVA O N, et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium[J]. Nature Communications, 2013, 4: 2785. |
| 98 | NAIZABEKOV S, LEE E Y. Genome-scale metabolic model reconstruction and in silico investigations of methane metabolism in Methylosinus trichosporium OB3b[J]. Microorganisms, 2020, 8(3):437. |
| 99 | 高子熹, 郭树奇, 费强. 生物转化温室气体生产单细胞蛋白的研究进展[J]. 化工学报,2021, 72(6): 3202-3214. |
| GAO Z X, GUO S Q, FEI Q. Recent progress in microbial bioconversion of greenhouse gases into single cell protein[J]. CIESC Journal,2021, 72(6): 3202-3214. | |
| 100 | PICONE N, MOHAMMADI S S, WAAJEN A C, et al. More than a methanotroph: a broader substrate spectrum for Methylacidiphilum fumariolicum SolV[J]. Frontiers in Microbiology, 2020, 11: 604485. |
| 101 | REN J, LEE H M, THAI T D, et al. Identification of a cytosine methyltransferase that improves transformation efficiency in Methylomonas sp. DH-1[J]. Biotechnology for Biofuels, 2020, 13(1): 200. |
| [1] | YING Hanjie, LIU Dong, WANG Zhenyu, SHEN Tao, ZHUANG Wei, ZHU Chenjie. Exploring industrial biomanufacturing and the goal of “carbon neutrality” [J]. Synthetic Biology Journal, 2025, 6(1): 1-7. |
| [2] | ZHANG Chenyue, MA Yingqun, WANG Xing, FU Rongzhan, HUANG Jiwei, HUA Xiufu, FAN Daidi, FEI Qiang. Progress in the bioconversion of biogas into sustainable aviation fuel [J]. Synthetic Biology Journal, 2023, 4(6): 1246-1258. |
| [3] | DIAO Zhidian, WANG Xixian, SUN Qing, XU Jian, MA Bo. Advances and applications of single-cell Raman spectroscopy testing and sorting equipment [J]. Synthetic Biology Journal, 2023, 4(5): 1020-1035. |
| [4] | WU Yujie, LIU Xinxin, LIU Jianhui, Yang Kaiguang, SUI Zhigang, ZHANG Lihua, ZHANG Yukui. Research progress of strain screening and quantitative analysis of key molecules based on high-throughput liquid chromatography and mass spectrometry [J]. Synthetic Biology Journal, 2023, 4(5): 1000-1019. |
| [5] | SUN Meili, WANG Kaifeng, LU Ran, JI Xiaojun. Rewiring and application of Yarrowia lipolytica chassis cell [J]. Synthetic Biology Journal, 2023, 4(4): 779-807. |
| [6] | GAO Xianyun, NIU Lingxue, JIAN Ni, GUAN Ningzi. Applications of microbial synthetic biology in the diagnosis and treatment of diseases [J]. Synthetic Biology Journal, 2023, 4(2): 263-282. |
| [7] | TU Ran, LI Shixin, LI Haoni, WANG Meng. Advances and applications of droplet-based microfluidics in evolution and screening of engineered microbial strains [J]. Synthetic Biology Journal, 2023, 4(1): 165-184. |
| [8] | WANG Xixian, SUN Qing, DIAO Zhidian, XU Jian, MA Bo. Advances with applications of Raman spectroscopy in single-cell phenotype sorting and analysis [J]. Synthetic Biology Journal, 2023, 4(1): 204-224. |
| [9] | LIU Qi, QIAN Zhilan, SONG Lili, YAO Chaoying, XU Mingqiang, REN Yanna, CAI Menghao. Rewiring and application of Pichia pastoris chassis cell [J]. Synthetic Biology Journal, 2022, 3(6): 1150-1173. |
| [10] | DONG Zhengxin, SUN Tao, CHEN Lei, ZHANG Weiwen. Applications of regulatory engineering in photosynthetic cyanobacteria [J]. Synthetic Biology Journal, 2022, 3(5): 966-984. |
| [11] | TAO Fei, SUN Tao, WANG Yu, WEI Ting, NI Jun, XU Ping. Challenges and opportunities in the research of Synechococcus chassis under the context of carbon peak and neutrality [J]. Synthetic Biology Journal, 2022, 3(5): 932-952. |
| [12] | CUI Jinyu, ZHANG Aidi, LUAN Guodong, LYU Xuefeng. Engineering microalgae for photosynthetic biosynthesis: progress and prospect [J]. Synthetic Biology Journal, 2022, 3(5): 884-900. |
| [13] | BI Jiacheng, TIAN Zhigang. Synthetic immunology and future NK cell immunotherapy [J]. Synthetic Biology Journal, 2022, 3(1): 22-34. |
| [14] | REN Shichao, SUN Qiuyan, FENG Xudong, LI Chun. Biosynthesis of pentacyclic triterpenoid saponins in microbial cell factories [J]. Synthetic Biology Journal, 2022, 3(1): 168-183. |
| [15] | CHEN Jiuzhou, WANG Yu, PU Wei, ZHENG Ping, SUN Jibin. Advances and perspective on bioproduction of 5-aminolevulinic acid [J]. Synthetic Biology Journal, 2021, 2(6): 1000-1016. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||