Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (3): 428-443.DOI: 10.12211/2096-8280.2021-023
Multiple interleaved RS codes for data storage using up to Mb-scale synthetic DNA in living cells
CHEN Weigang1,2, GE Qi1, WANG Panpan1, HAN Mingzhe2,3, GUO Jian1
- 1.School of Microelectronics,Tianjin University,Tianjin 300072,China
2.Frontiers Science Center for Synthetic Biology (MOE),Tianjin University,Tianjin 300072,China
3.School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
-
Received:2021-02-09Revised:2021-03-28Online:2021-07-14Published:2021-06-30 -
Contact:CHEN Weigang
细胞内大片段DNA数据存储的多RS码交织编码
陈为刚1,2, 葛奇1, 王盼盼1, 韩明哲2,3, 郭健1
- 1.天津大学微电子学院,天津 300072
2.教育部合成生物学前沿科学中心,天津大学,天津 300072
3.天津大学化工学院,天津 300072
-
通讯作者:陈为刚 -
作者简介:陈为刚 (1980—),男,博士,副教授。研究方向为DNA数据存储、信息论与编码理论。 E-mail:chenwg@tju.edu.cn
CLC Number:
Cite this article
CHEN Weigang, GE Qi, WANG Panpan, HAN Mingzhe, GUO Jian. Multiple interleaved RS codes for data storage using up to Mb-scale synthetic DNA in living cells[J]. Synthetic Biology Journal, 2021, 2(3): 428-443.
陈为刚, 葛奇, 王盼盼, 韩明哲, 郭健. 细胞内大片段DNA数据存储的多RS码交织编码[J]. 合成生物学, 2021, 2(3): 428-443.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-023
| 主要结果 | 总体逻辑密度 (包含引物或载体骨架) /bit·bp-1 | 数据部分逻辑密度(不包含引物或载体骨架) /bit·bp-1 |
|---|---|---|
| Church等[ | 0.60 | 0.83 |
| Goldman等[ | 0.19 | 0.29 |
| Grass等[ | 0.83 | 1.16 |
| Bornholt等[ | 0.57 | 0.85 |
| Erlich等[ | 1.19 | 1.57 |
| Blawat等[ | 0.89 | 1.08 |
| Organick等[ | 0.81 | 1.10 |
| Chen 等[ | 1.19 | 1.24 |
| Ping 等[ | 1.32 | 1.88 |
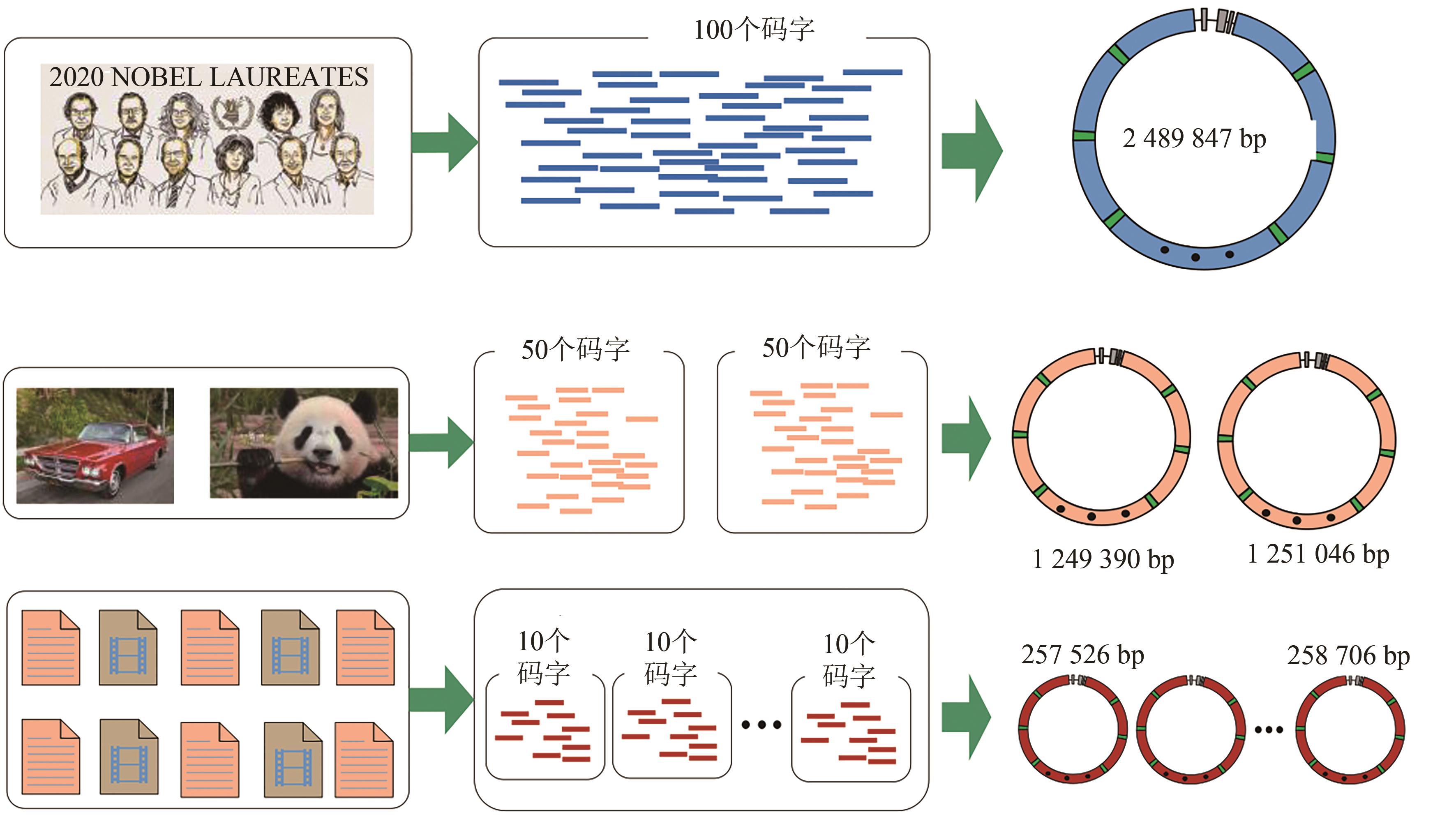
| 本工作(单段DNA,2 489 847 p) | 1.947 | 1.973 |
Tab. 1 Base logical density using different encoding schemes
| 主要结果 | 总体逻辑密度 (包含引物或载体骨架) /bit·bp-1 | 数据部分逻辑密度(不包含引物或载体骨架) /bit·bp-1 |
|---|---|---|
| Church等[ | 0.60 | 0.83 |
| Goldman等[ | 0.19 | 0.29 |
| Grass等[ | 0.83 | 1.16 |
| Bornholt等[ | 0.57 | 0.85 |
| Erlich等[ | 1.19 | 1.57 |
| Blawat等[ | 0.89 | 1.08 |
| Organick等[ | 0.81 | 1.10 |
| Chen 等[ | 1.19 | 1.24 |
| Ping 等[ | 1.32 | 1.88 |
| 本工作(单段DNA,2 489 847 p) | 1.947 | 1.973 |
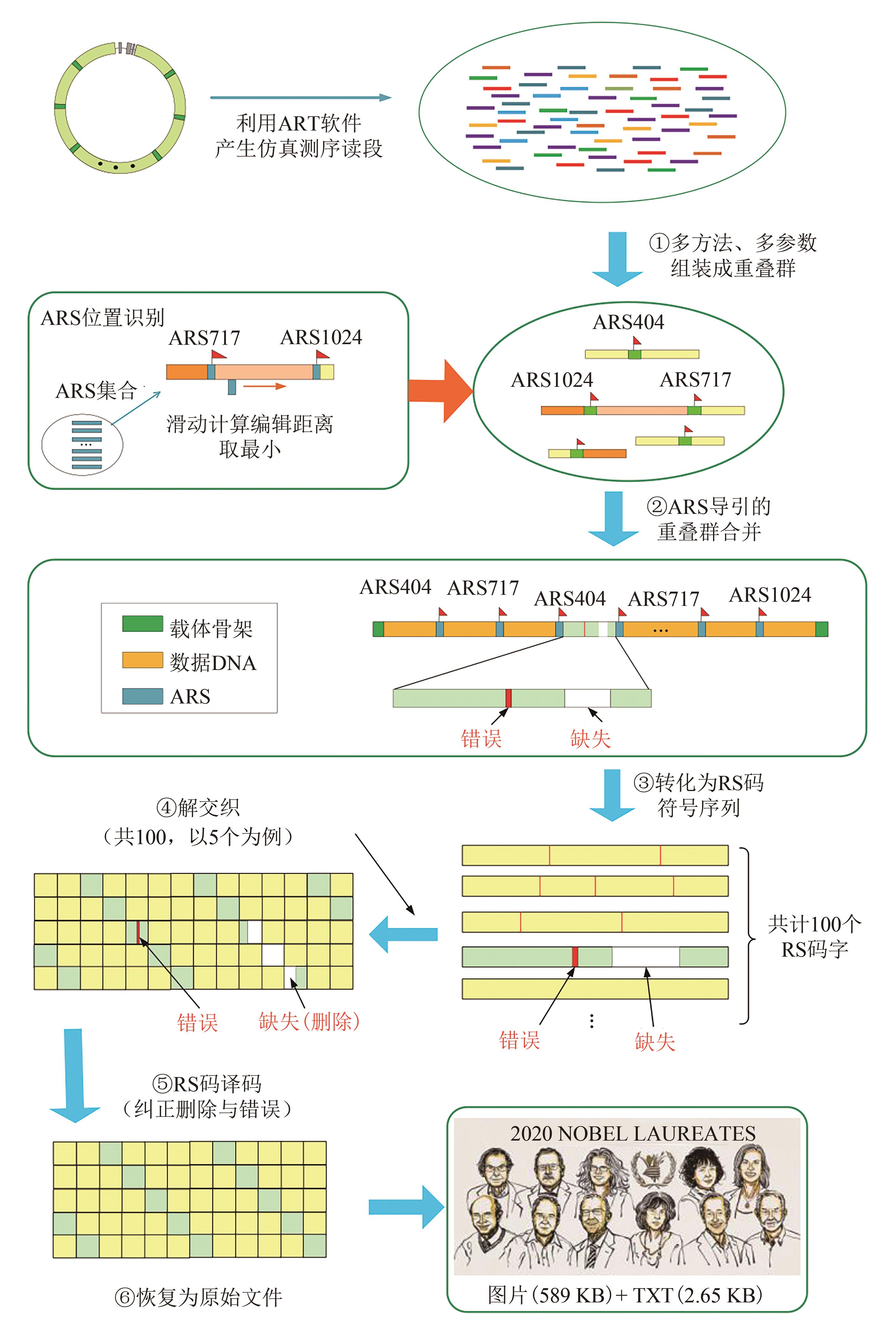
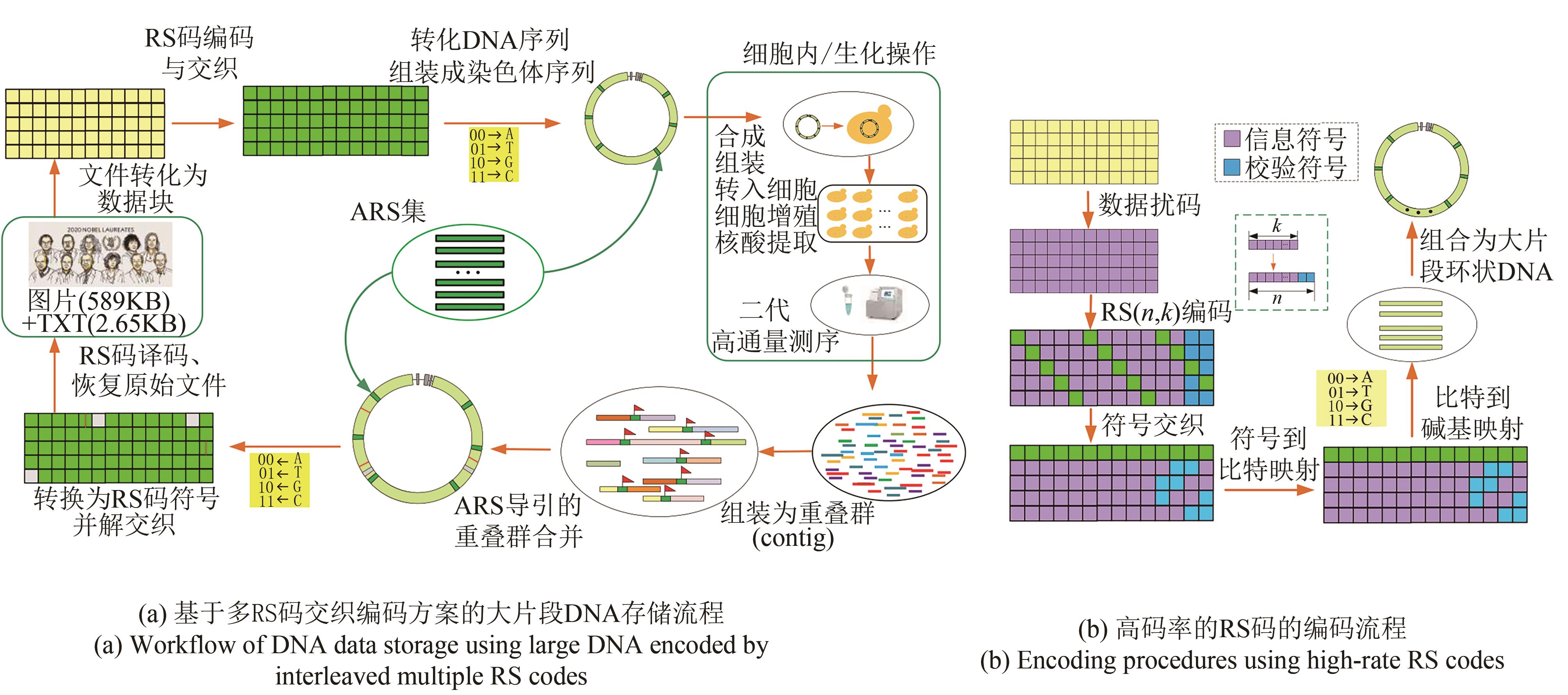
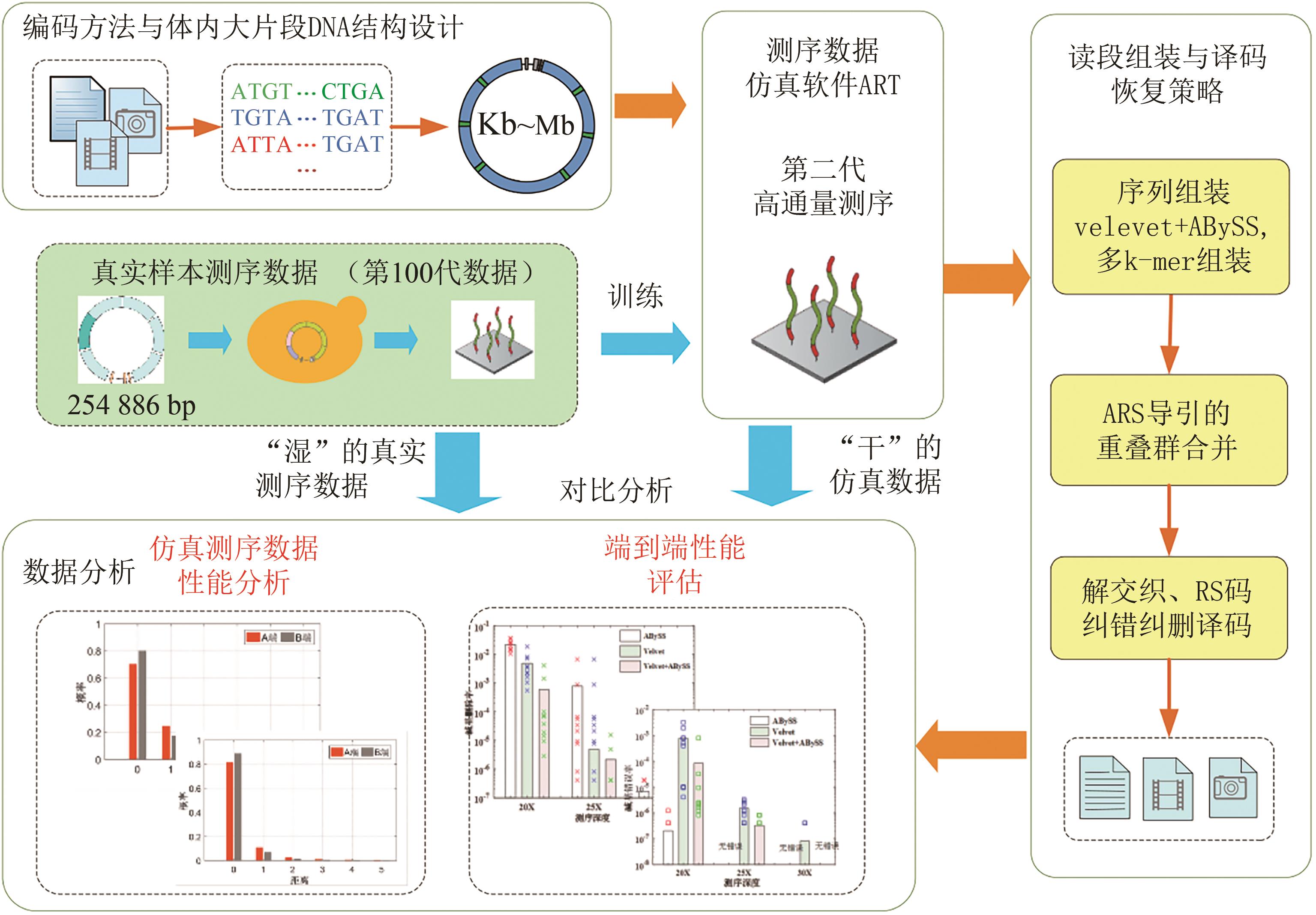
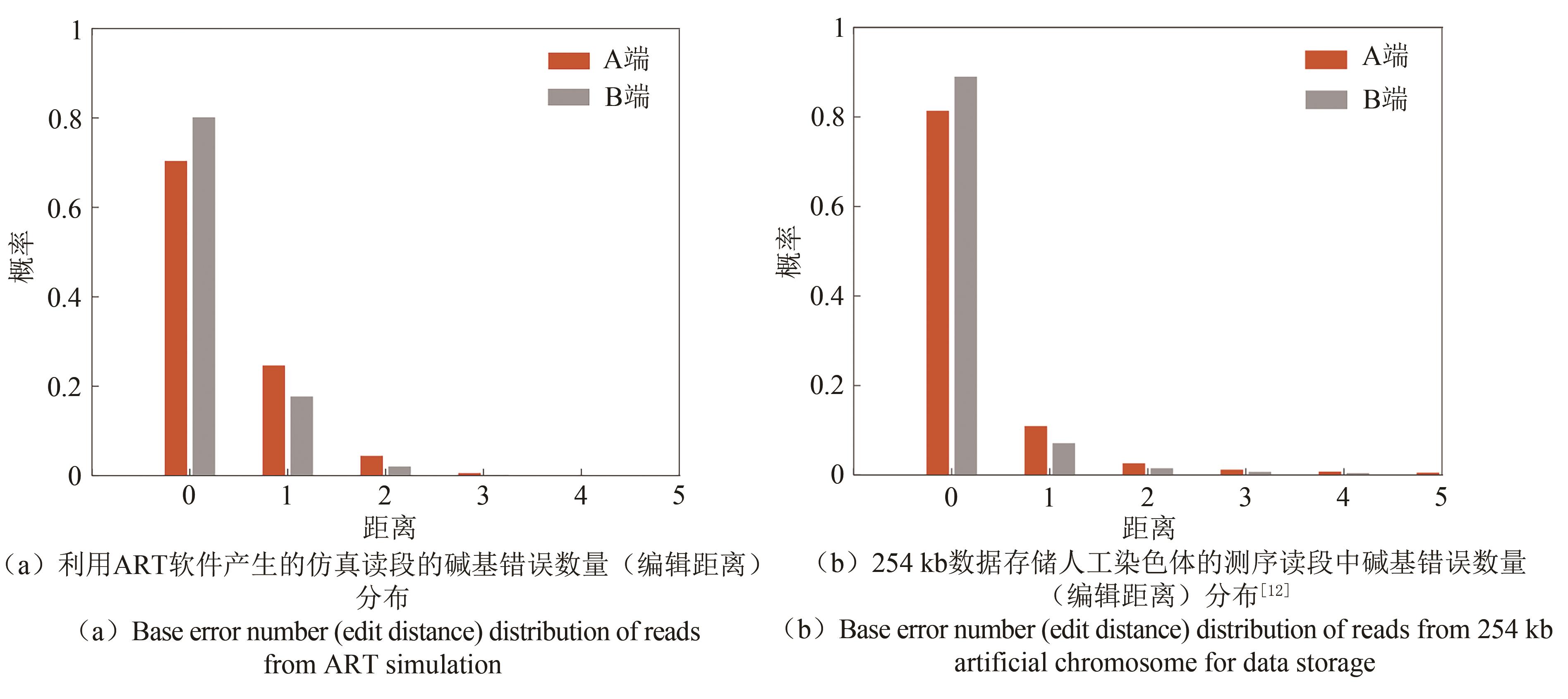
Fig. 3 Data readout processes (The process includes de novo assembly from short reads using multiple programs with multiple k-mers, ARS navigated combination of multiple contigs, converting into symbol sequences of RS code, deinterleaving, and RS erasure and error correction)
测序 覆盖度 | 组装软件 | k-mer | 非交织RS码不能恢复数据段数量以及缺失与错误(缺失数量+错误数量) | 解交织后RS码译码 |
|---|---|---|---|---|
| 20× | Velvet | {25,27,29,31,33} | 实验1:2个(2705+0,2450+7711) | 成功 |
| 实验2:3个(3525+0,673+1913,5194+0) | 成功 | |||
| 实验3:5个(6779+0,11003+0, 45+1855,9296+0,18612+0) | 失败 | |||
| 实验4:1个(5498+0) | 成功 | |||
| 实验5:1个(17306+0) | 成功 | |||
| 实验6:1个(8387+0) | 成功 | |||
| 实验7:1个(1134+0) | 成功 | |||
| 实验8:3个(2668+16,8320+0,9298+919) | 成功 | |||
| 实验9:1个(2466+1213) | 成功 | |||
| 实验10:2个(3657+0,0+4712) | 成功 | |||
| Velvet+ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验2(3525+1,1+1905) | 成功 | |
| 实验5(10041+0) | 成功 | |||
| 25× | Velvet | 25 | 实验6(937+0) | 成功 |
| 实验9(10620+1) | 成功 | |||
| 27 | 实验2(13604+1) | 成功 | ||
| 29 | 实验8(21488+0,3373+0) | 成功 | ||
| 实验10(13023+17) | 成功 | |||
| 31 | 实验2(1692+0) | 成功 | ||
| 实验7(1105+0) | 成功 | |||
| 实验8(3373+0) | 成功 | |||
| 33 | 实验2(1696+0) | 成功 | ||
| 实验10(1854+1986) | 成功 | |||
| {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | ||
| ABySS | {25,27,29,31,33} | 实验1(16950+0) | 成功 | |
| 实验8(2151+0) | 成功 | |||
| Velvet+ ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | |
| 30× | Velvet | 27 | 实验1~7, 9,10均无大片段错误 实验8(1292+0) | 成功 |
| {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | ||
| ABySS | {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | |
| Velvet+ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 |
Tab. 2 Data recovery analysis using interleaved multiple RS codes
测序 覆盖度 | 组装软件 | k-mer | 非交织RS码不能恢复数据段数量以及缺失与错误(缺失数量+错误数量) | 解交织后RS码译码 |
|---|---|---|---|---|
| 20× | Velvet | {25,27,29,31,33} | 实验1:2个(2705+0,2450+7711) | 成功 |
| 实验2:3个(3525+0,673+1913,5194+0) | 成功 | |||
| 实验3:5个(6779+0,11003+0, 45+1855,9296+0,18612+0) | 失败 | |||
| 实验4:1个(5498+0) | 成功 | |||
| 实验5:1个(17306+0) | 成功 | |||
| 实验6:1个(8387+0) | 成功 | |||
| 实验7:1个(1134+0) | 成功 | |||
| 实验8:3个(2668+16,8320+0,9298+919) | 成功 | |||
| 实验9:1个(2466+1213) | 成功 | |||
| 实验10:2个(3657+0,0+4712) | 成功 | |||
| Velvet+ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验2(3525+1,1+1905) | 成功 | |
| 实验5(10041+0) | 成功 | |||
| 25× | Velvet | 25 | 实验6(937+0) | 成功 |
| 实验9(10620+1) | 成功 | |||
| 27 | 实验2(13604+1) | 成功 | ||
| 29 | 实验8(21488+0,3373+0) | 成功 | ||
| 实验10(13023+17) | 成功 | |||
| 31 | 实验2(1692+0) | 成功 | ||
| 实验7(1105+0) | 成功 | |||
| 实验8(3373+0) | 成功 | |||
| 33 | 实验2(1696+0) | 成功 | ||
| 实验10(1854+1986) | 成功 | |||
| {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | ||
| ABySS | {25,27,29,31,33} | 实验1(16950+0) | 成功 | |
| 实验8(2151+0) | 成功 | |||
| Velvet+ ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | |
| 30× | Velvet | 27 | 实验1~7, 9,10均无大片段错误 实验8(1292+0) | 成功 |
| {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | ||
| ABySS | {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | |
| Velvet+ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 |
| 1 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA [J]. Science, 2012, 337(6102): 1628. |
| 2 | GOLDMAN N, BERTONE P, CHEN S Y, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA [J]. Nature, 2013, 494(7435): 77-80. |
| 3 | MEISER L C, ANTKOWIAK P L, KOCH J, et al. Reading and writing digital data in DNA [J]. Nature Protocols, 2020, 15(1): 86-101. |
| 4 | DONG Y M, SUN F J, PING Z, et al. DNA storage: research landscape and future prospects [J]. National Science Review, 2020, 7(6): 1092-1107. |
| 5 | PING Z, MA D Z, HUANG X L, et al. Carbon-based archiving: current progress and future prospects of DNA-based data storage [J]. Gigascience, 2019, 8(6): giz075. |
| 6 | 董一名, 孙法家, 武瑞君, 等. DNA数字信息存储的研究进展[J]. 合成生物学, 2021, 2(3): 323-334. |
| DONG Yiming, SUN Fajia, WU Ruijun, et al. Research progress on DNA molecules for digital information storage [J]. Synthetic Biology Journal, 2021, 2(3): 323-334. | |
| 7 | 韩明哲,陈为刚, 宋理富, 等. DNA信息存储:生命系统与信息系统的桥梁 [J]. 合成生物学, 2021.2(3): 309-322. |
| HAN Mingzhe, CHEN Weigang, SONG Lifu,et al. DNA information storage:bridging biological and digital world [J]. Synthetic Biology Journal, 2021, 2(3):309-322. | |
| 8 | Semiconductor Industry Association (ISA), Semiconductor Research Corporation (SRC). The decadal plan for semiconductors [R/OL]. 2021. . |
| 9 | BORNHOLT J, LOPEZ R, CARMEAN D M, et al. Toward a DNA-based archival storage system [J]. IEEE Micro, 2017, 37(3): 98-104. |
| 10 | ERLICH Y, ZIELINSKI D. DNA fountain enables a robust and efficient storage architecture [J]. Science, 2017, 355(6328): 950-954. |
| 11 | YAZDI TABATABAEI HOSSEIN S M, GABRYS R, MILENKOVIC O. Portable and error-free DNA-based data storage [J]. Scientific Reports, 2017, 7(1): 5011. |
| 12 | ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage [J]. Nature Biotechnology, 2018, 36(3): 242-248. |
| 13 | GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes [J]. Angewandte Chemie International Edition, 2015, 54(8): 2552-2555. |
| 14 | BLAWAT M, GAEDKE K, HUETTER I, et al. Forward error correction for DNA data storage [J]. Procedia Computer Science, 2016, 80: 1011-1022. |
| 15 | 陈为刚, 黄刚, 李炳志, 等. 音视频文件的DNA信息存储 [J]. 中国科学: 生命科学, 2020, 50(1): 81-85. |
| CHEN Weigang, HUANG Gang, LI Bingzhi, et al. DNA information storage for audio and video files [J]. SCIENTIA SINICA Vitae, 2020, 50(1): 81-85. | |
| 16 | PING Z, CHEN S, HUANG X, et al. Towards practical and robust DNA-based data archiving by codec system named Yin-Yang [EB/OL]. [2021-05-27]. . |
| 17 | PRESS W H, HAWKINS J A, JONES S K, et al. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints [J]. Proceedings of the National Academy of Science of United States of America, 2020, 117(31): 18489-18496. |
| 18 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications [J]. Nature Methods, 2014, 11(5): 499-507. |
| 19 | CHEN W G, HAN M Z, ZHOU J T, et al. An artificial chromosome for data storage, [J] National Science Review, 2021, 8(5): nwab028. |
| 20 | DAVIS J. Microvenus [J]. Art Journal, 1996, 55(1): 70-74. |
| 21 | SHIPMAN S L, NIVALA J, MACKLIS J D, et al. CRISPR–Cas encoding of a digital movie into the genomes of a population of living bacteria [J]. Nature, 2017, 547(7663): 345-349. |
| 22 | HAO M, QIAO H Y, GAO Y, et al. A mixed culture of bacterial cells enables an economic DNA storage on a large scale [J]. Communications Biology, 2020, 3(1): 416-424. |
| 23 | NGUYEN H H, PARK J, PARK S J, et al. Long-term stability and integrity of plasmid-based DNA data storage [J]. Polymers, 2018, 10(1): 28. |
| 24 | YIM S S, MCBEE R M, SONG A M, et al. Robust direct digital-to-biological data storage in living cells [J]. Nature Chemical Biology, 2021,17(3):246-253. |
| 25 | POSTMA E D, DASHKO S, BREEMEN L V, et al. A supernumerary designer chromosome for modular in vivo pathway assembly in Saccharomyces cerevisiae [J]. Nucleic Acids Research, 2021, 49(3): 1769-1783. |
| 26 | SONG L F, ZENG A P. Orthogonal information encoding in living cells with high error-tolerance, safety, and fidelity [J]. ACS Synthetic Biology, 2018, 7(3): 866-874. |
| 27 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome [J]. Science, 2010, 329(5987): 52-56. |
| 28 | WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X [J]. Science, 2017, 355(6329): eaaf4706. |
| 29 | XIE Z X, LI B Z, MITCHELL L A, et al. "Perfect" designer chromosome V and behavior of a ring derivative [J]. Science, 2017, 355(6329): eaaf4704. |
| 30 | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome [J]. Science, 2017, 355(6329): eaaf4791. |
| 31 | FREDENS J, WANG K H, TORRE D D L, et al. Total synthesis of Escherichia coli with a recoded genome [J]. Nature, 2019, 569(7757): 514-518. |
| 32 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING Mingzhu, LI Bingzhi, WANG Ying, et al. Significant research progress in synthetic biology [J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 33 | 彭凯, 逯晓云, 程健, 等. DNA合成、组装与纠错技术研究进展[J]. 合成生物学, 2020, 1(6): 697-708. |
| PENG Kai, LU Xiaoyun, CHENG Jian, et al. Advances in technologies for de novo DNA synthesis,assembly and error correction [J]. Synthetic Biology Journal, 2020, 1(6): 697-708. | |
| 34 | 刘晓, 王慧媛, 熊燕, 等. 基因合成与基因组编辑[J]. 中国细胞生物学学报, 2019, 41(11): 2072-2082. |
| LIU Xiao, WANG Huiyuan, XIONG Yan, et al. Progress in gene synthesis and genome editing [J]. Chinese Journal of Cell Biology, 2019, 41(11): 2072-2082. | |
| 35 | 罗周卿, 戴俊彪. 合成基因组学:设计与合成的艺术[J]. 生物工程学报, 2017, 33(3): 331-342. |
| LUO Zhouqing, DAI Junbiao. Synthetic genomics: the art of design and synthesis [J]. Chinese Journal of Biotechnology 2017, 33(3): 331-342. | |
| 36 | 王会, 戴俊彪, 罗周卿. 基因组的"读-改-写"技术[J]. 合成生物学, 2020, 1(5): 503-515. |
| WANG Hui, DAI Junbiao, LUO Zhouqing. Reading, editing, and writing techniques for genome research [J]. Synthetic Biology Journal, 2020, 1(5): 503-515. | |
| 37 | KARAS B J, JABLANOVIC J, SUN L J, et al. Direct transfer of whole genomes from bacteria to yeast [J]. Nature Methods, 2013, 10(5): 410-412. |
| 38 | 朱雪龙. 应用信息论基础[M]. 北京: 清华大学出版社, 2001. |
| ZHU X L.Fundamentals of applied information theory[M]. Beijing: Tsinghua University Press, 2001. | |
| 39 | LIN S, COSTELLO D J. Error control coding [M]. London:Pearson Education Inc, 2004. |
| 40 | REED I S, SOLOMON G. Polynomial codes over certain finite fields [J]. Journal of the Society for Industrial & Applied Mathematics, 1960, 8(2): 300-304. |
| 41 | MATSUI H, MITA S. A new encoding and decoding system of Reed-Solomon codes for HDD [J]. IEEE Transactions on Magnetics, 2009, 45(10): 3757-3760. |
| 42 | RIGGLE C M, MCCARTHY S G. Design of error correction systems for disk drives [J]. IEEE Transactions on Magnetics, 1998, 34(4): 2362-2371. |
| 43 | LEE Joohyun, LEE Jaejin, PARK T. Error control scheme for high-speed DVD systems [J]. IEEE Transactions on Consumer Electronics, 2005, 51(4): 1197-1203. |
| 44 | SONG M A, KUO S Y, LAN I F. A low complexity design of Reed Solomon code algorithm for advanced RAID system [J]. IEEE Transactions on Consumer Electronics, 2007, 53(2): 265-273. |
| 45 | IM S, SHIN D. Flash-Aware RAID techniques for dependable and High-Performance flash memory SSD [J]. Computers IEEE Transactions on Computers, 2011, 60(1): 80-92. |
| 46 | HUANG J Z, LIANG X H, QIN X, et al. Scale-RS: an efficient scaling scheme for RS-Coded storage clusters [J]. IEEE Transactions on Parallel & Distributed Systems, 2015, 26(6): 1704-1717. |
| 47 | CHEN W G, WANG T, HAN C C, et al. Erasure-correction-enhanced iterative decoding for LDPC-RS product codes [J]. China Communications, 2021, 18(1): 49-60. |
| 48 | SIOW C C, NIEDUSZYNSKA S R, MÜLLER C A, et al. OriDB, the DNA replication origin database updated and extended [J]. Nucleic Acids Research, 2012, 40(D1): 682-686. |
| 49 | LOMAN N J, MISRA R V, DALLMAN T J, et al. Performance comparison of benchtop high-throughput sequencing platforms [J]. Nature Biotechnology, 2012, 30(5): 434-439. |
| 50 | MARDIS E R. Next-generation DNA sequencing methods [J]. Annual Review of Genomics and Human Genetics, 2008, 9(1): 387-402. |
| 51 | SHENDURE J, JI H. Next-generation DNA sequencing [J]. Nature Biotechnology, 2008, 26(10): 1135-1145. |
| 52 | METZKER M L. Sequencing technologies—the next generation [J]. Nature Reviews Genetics, 2010, 11(1): 31-46. |
| 53 | COMPEAU P E C, PEVZNER P A, TESLER G. How to apply de Bruijn graphs to genome assembly [J]. Nature Biotechnology, 2011, 29(11): 987. |
| 54 | ZERBINO D R, BIRNEY E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs [J]. Genome Research, 2008, 18(5): 821-829. |
| 55 | SIMPSON J T, WONG K, JACKMAN S D, et al. ABySS: a parallel assembler for short read sequence data [J]. Genome Research, 2009, 19(6): 1117-1123. |
| 56 | HUANG W C, LI L P, MYERS J R, et al. ART: a next-generation sequencing read simulator [J]. Bioinformatics, 2012, 28: 593-594. |
| 57 | CHEN W G, WANG L X, HAN M Z, et al. Sequencing barcode construction and identification methods based on block error-correction codes [J]. Science China Life Sciences, 2020,63(10):1580-1592. |
| 58 | CHEN W G, WANG P P, WANG L X, et al. Low-complexity and highly robust barcodes for error-rich single molecular sequencing [J]. 3 Biotech, 2021, 11(2): 1-11. |
| 59 | 格林 M R, 萨姆布鲁克 J. 分子克隆实验指南 [M]. 贺福初, 陈薇, 杨晓明, 译. 4版. 北京: 科学出版社, 2013: 226-227. |
| GREEN M R, SAMBROOK J. Molecular cloning: a laboratory manual [M]. HE F C, CHEN W, YANG X M, trans. 4th ed. Beijing: Science Press, 2013: 226-227. | |
| 60 | DUNNEN J D, GROOTSCHOLTEN P M, DAUWERSE J, et al. Reconstruction of the 2.4 Mb human DMD-gene by homologous YAC recombination [J]. Human Molecular Genetics, 1992, 1(1): 19-28. |
| [1] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [2] | HUANG Xiaoluo, DAI Junbiao. DNA synthesis technology: foundation of DNA data storage [J]. Synthetic Biology Journal, 2021, 2(3): 335-353. |
| [3] | GAO Yanmin, TANG Mengtong, LIU Qian, QIAO Hongyan, WANG Taoxue, QI Hao. The pivotal biochemical methods in DNA data storage [J]. Synthetic Biology Journal, 2021, 2(3): 384-398. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||