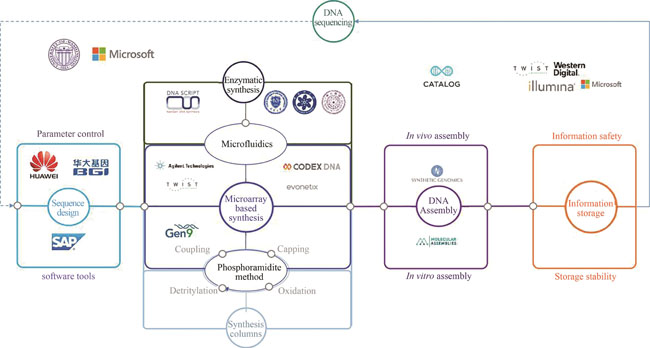
With the rapid growth of digital data production, there is a strong motivation for developing new data storage media. DNA, as a medium for data storage, has attracted the attention of many research institutions and enterprises. DNA based storage technology is expeditiously evolving and has great market potential. It is predicted that, by 2024, about 30% of digital businesses will begin to try to use DNA for data storage. To meet the huge market demand, a matching patent scheme has become extraordinarily important for winning the initiative in the competition. This article analyzes the development trend of DNA synthesis and storage technology from the perspective of patent analysis. We have searched and obtained 1833 patents related to DNA synthesis and storage on a global scale (excluding gene sequencing patents required for DNA storage technology, also excluding DNA synthesis patents dedicated to diagnosis, treatment and other applications) by comprehensively using keywords, international patent classification, patentee, inventor search and other methods. Based on individual reading and comparison, we have utilized patent value analysis, citation analysis, cluster analysis, technical efficacy analysis and other methods to select representative patents that are expected to provide references for further investigations, patent layout and operation decisions in this field. To achieve the goal of using DNA to store digital information, the synthesis of oligonucleotides or polynucleotides is the basis of "writing". So far, the global oligonucleotide or polynucleotide synthesis technology has experienced three generations of development. The first and second generations use phosphoramidite chemical synthesis method, while the third generation is based on the principle of enzymatic synthesis. Global patent analysis has revealed that the number of patents published each year greatly increases while the synthesis technology of oligonucleotides or polynucleotides evolves to phosphoramidite chemical with combination of microarray-based chip technology. Many established companies have joined the development of the second generation of synthetic technology. The emergence of the third generation synthesis technology, which is based on the use of polymerases such as terminal deoxyribonucleoside transferase, is also reflected by the number of patents. Meanwhile, techniques required for DNA based storage, such as nucleic acid assembly, sequence design, and information storage, are also rapidly developing. The patentees corresponding to these technologies have shown obvious cross-industry integration features, since companies such as Microsoft, Intel, and Huawei have successively joined the competition and cooperation of DNA storage patents. It is foreseeable that in the future DNA based storage technology will inevitably involve the convergence of technologies in multiple fields such as Chemistry, Materials, Biology, Informatics, Mechanics, and Electronics. The further integration of these technologies would promote a regularly upgrading development path similar to "Moore's Law" in the field of DNA storage. Based on high-throughput, high-efficiency, high-fidelity and low-cost DNA synthesis, using comprehensive information encoding and decoding, integrating "edit"-"write"-"read"-"dissolve" functions, DNA storage system in the future will become a truly "usable" solution.