Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (3): 354-370.DOI: 10.12211/2096-8280.2020-089
• Invited Review • Previous Articles Next Articles
In situ chemical synthesis of DNA microarrays
YAN Han, XIAO Pengfeng, LIU Quanjun, LU Zuhong
- State Key Laboratory of Bioelectronics,School of Biological Sciences and Biomedical Engineering,Southeast University,Nanjing 210096,Jiangsu,China
-
Received:2020-12-21Revised:2021-04-09Online:2021-07-13Published:2021-06-30 -
Contact:LIU Quanjun, LU Zuhong
DNA微阵列原位化学合成
闫汉, 肖鹏峰, 刘全俊, 陆祖宏
- 东南大学生物科学与生物医学工程学院,生物电子学国家重点实验室,江苏 南京 210096
-
通讯作者:刘全俊,陆祖宏 -
作者简介:闫汉 (1991—),男,博士研究生。研究方向为DNA合成自组装,纳米孔生物分子检测。E-mail:230209118@seu.deu.cn刘全俊 (1968—),男,教授,博士生导师。研究方向为新一代基因测序、基因芯片、生物与化学传感器、单分子检测。E-mail:lqj@seu.edu.cn陆祖宏 (1960—),男,教授,博士生导师。研究方向为生物电子学,超分子组装、生物传感器、生物信息技术等。 E-mail:zhl@seu.deu.cn -
基金资助:国家自然科学基金(60121101)
CLC Number:
Cite this article
YAN Han, XIAO Pengfeng, LIU Quanjun, LU Zuhong. In situ chemical synthesis of DNA microarrays[J]. Synthetic Biology Journal, 2021, 2(3): 354-370.
闫汉, 肖鹏峰, 刘全俊, 陆祖宏. DNA微阵列原位化学合成[J]. 合成生物学, 2021, 2(3): 354-370.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-089
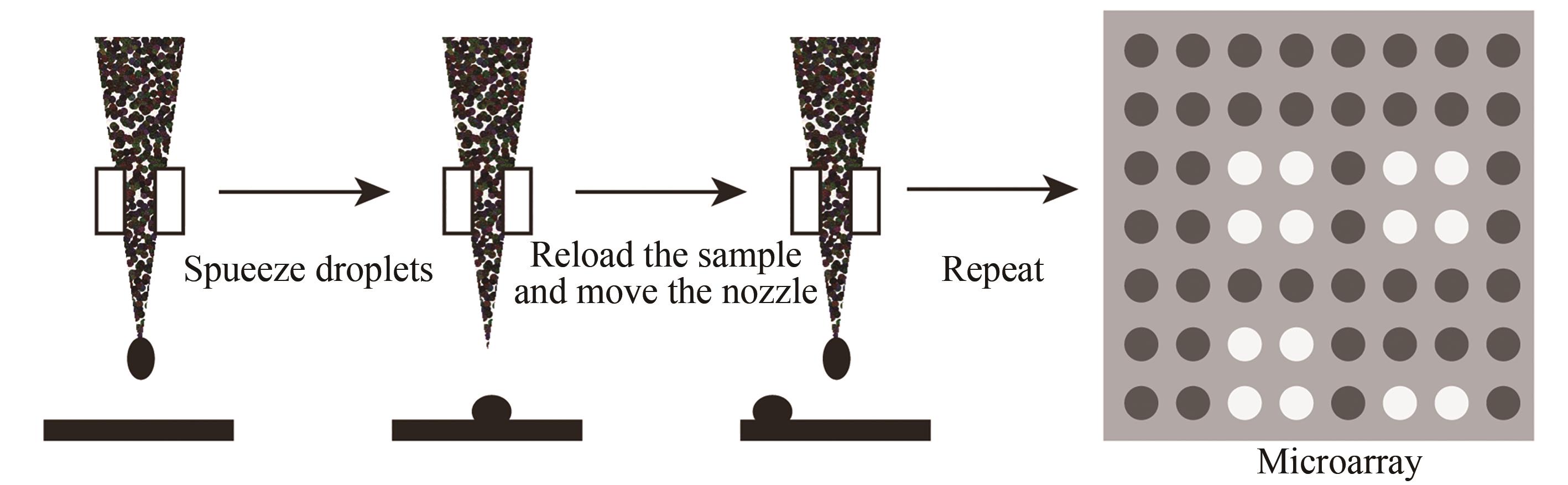
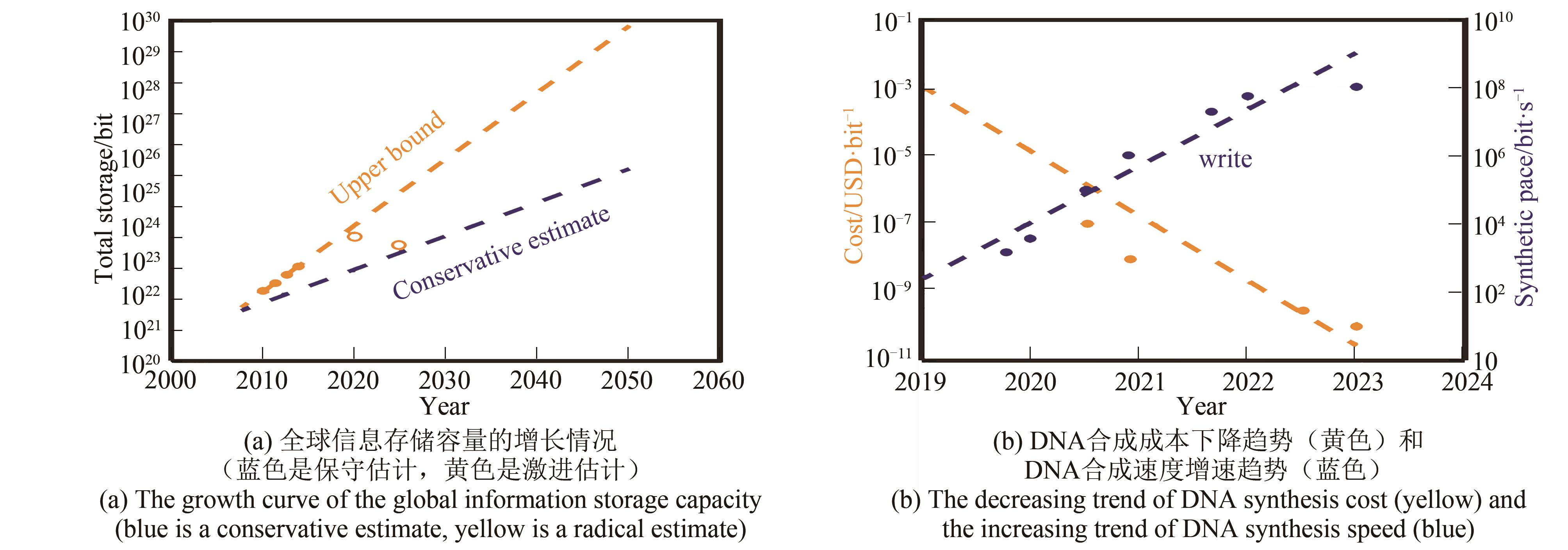
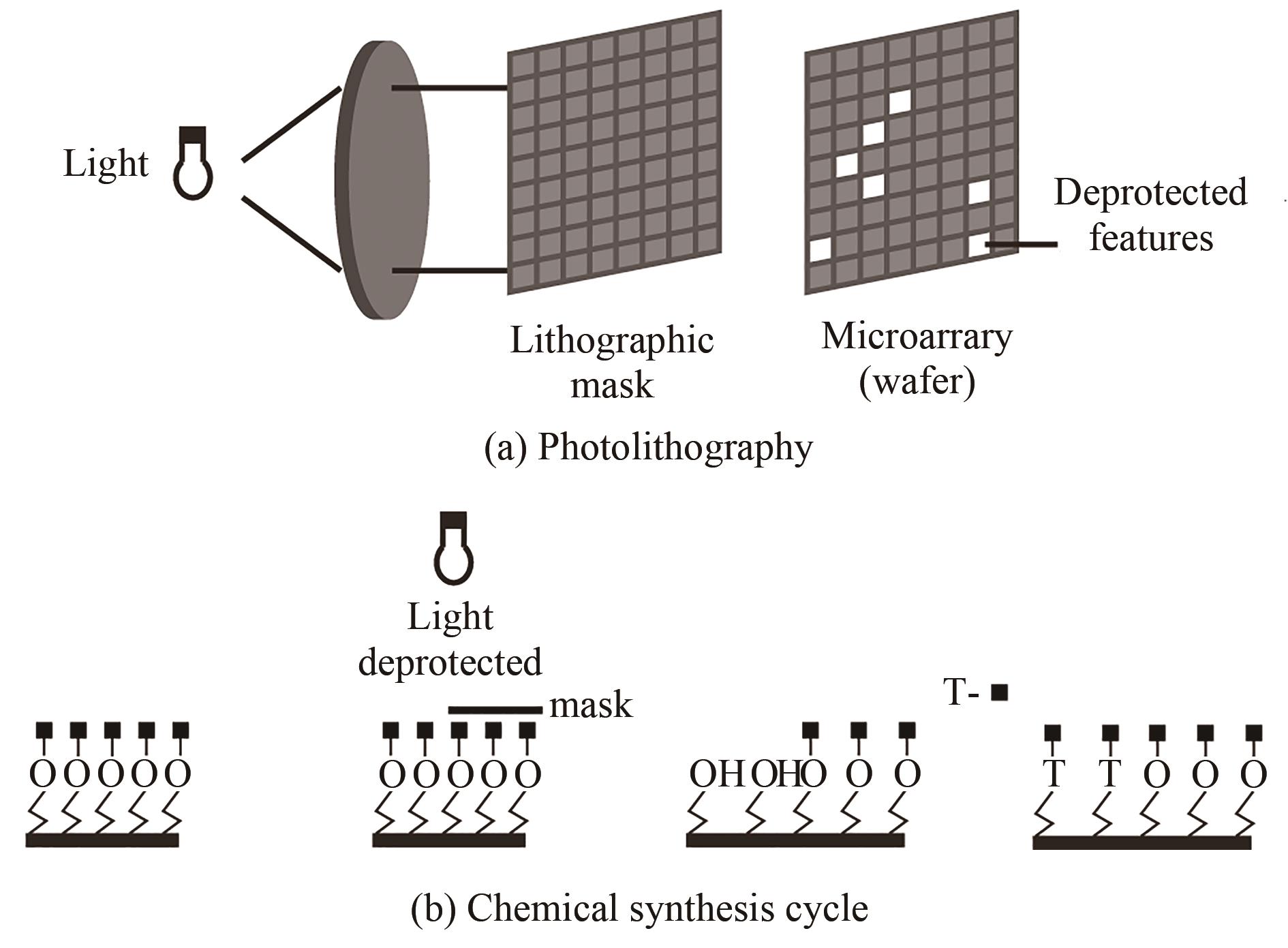
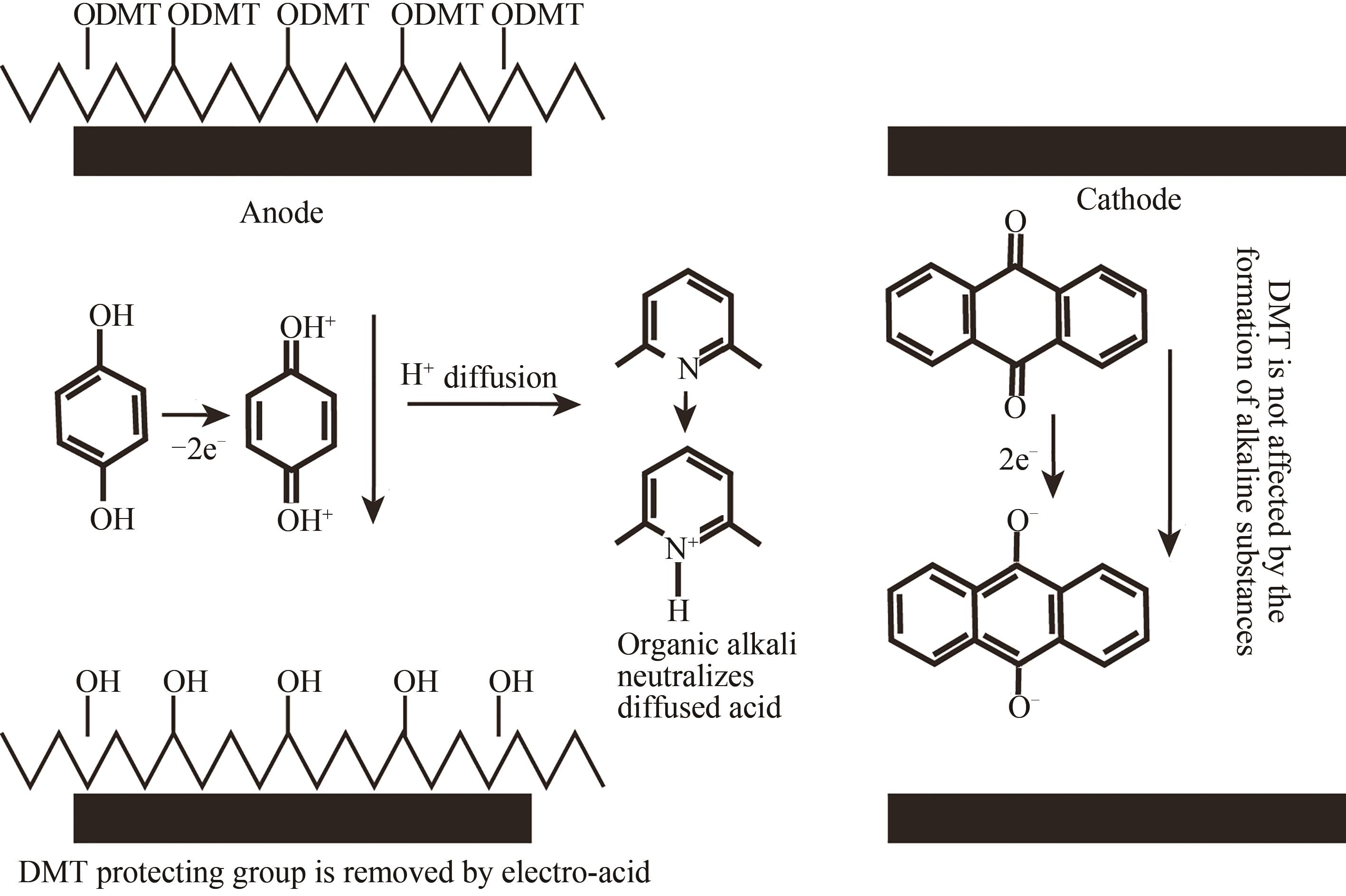
Fig. 8 Schematic diagram of the in-situ printing synthesis method(The four base synthesis reagents in the "cartridge" are respectively spotted to the predetermined position of the substrate to synthesize the base, and this step can be cycled to synthesize a preset DNA sequence in different positions of the substrate)
| 方法 | 技术特征 | 优点 | 缺点 |
|---|---|---|---|
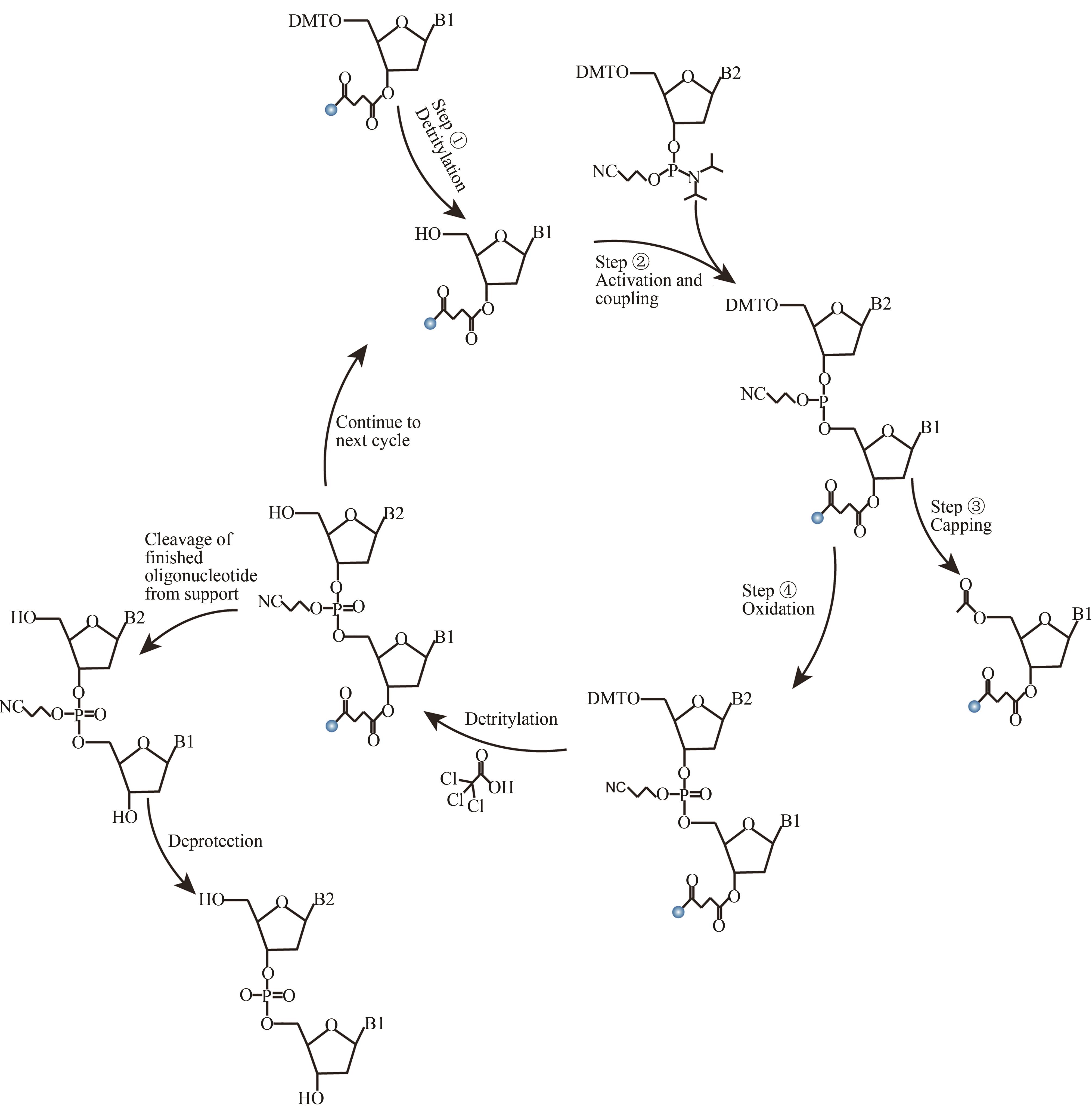
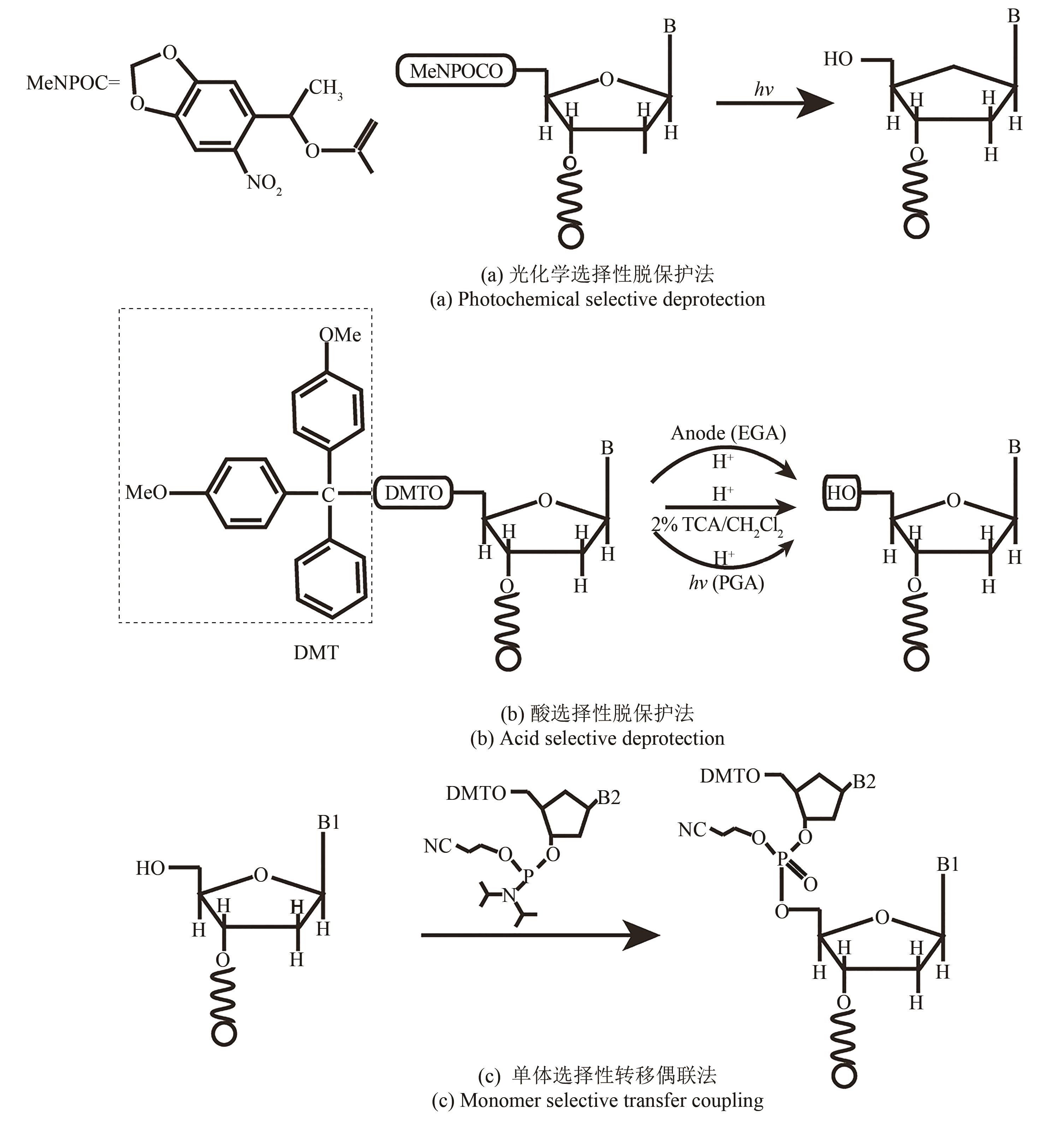
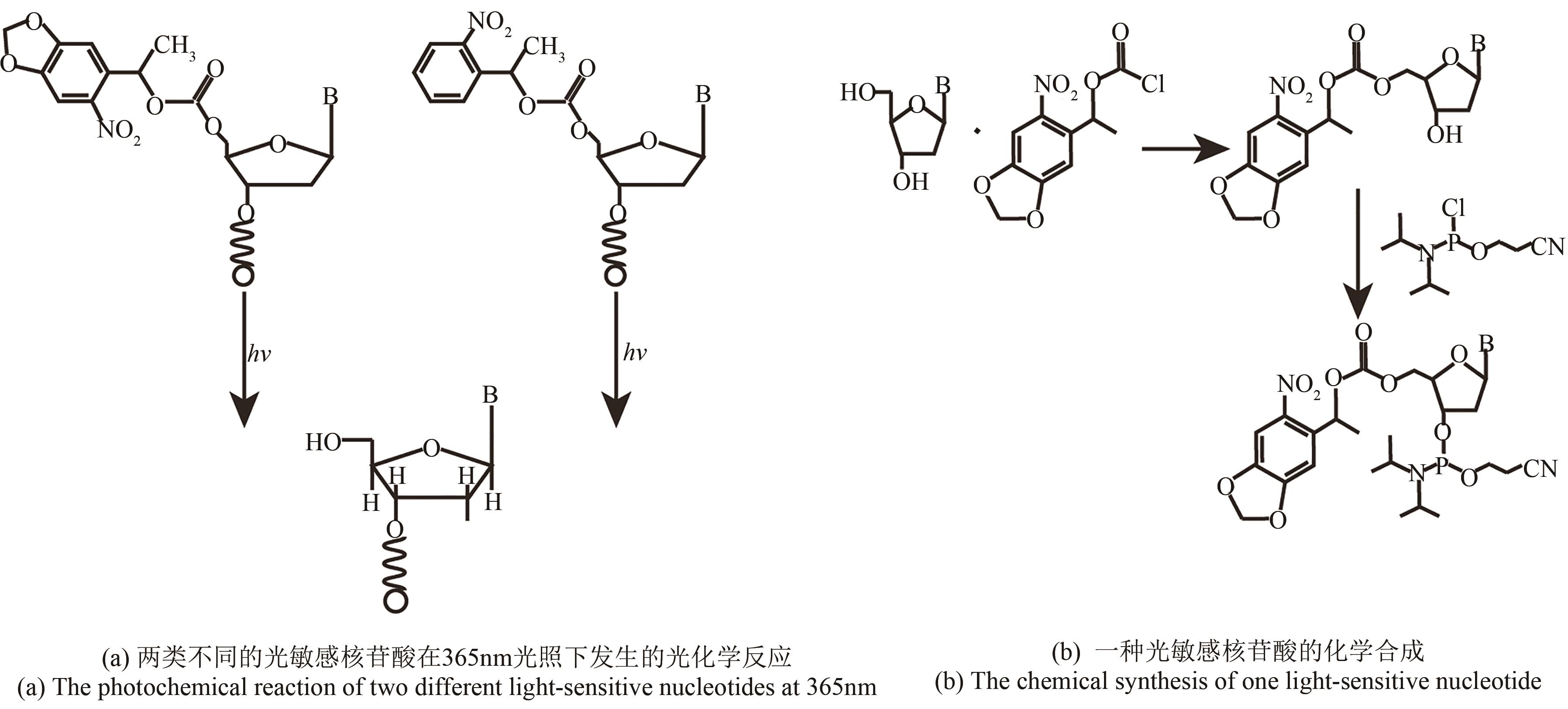
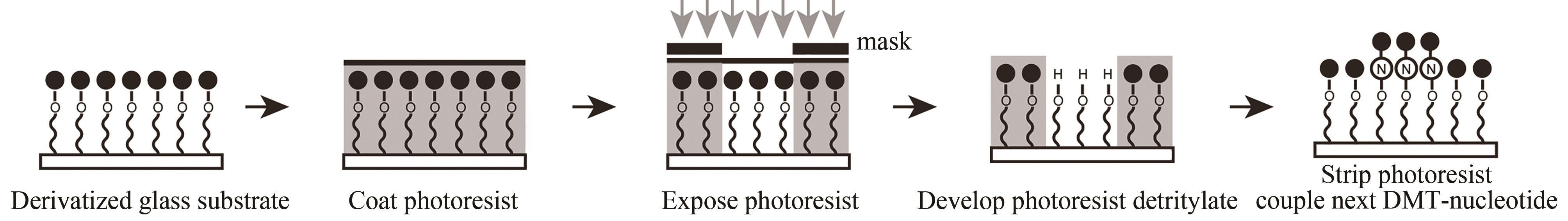
| 原位光刻法 | 光通过掩模(或者虚拟掩模)照射特定的阵点,5'端保护基团发生光化学反应被直接脱除,被活化的5'-羟基可以进行偶联、封闭、氧化反应 | 光刻的分辨率高。可合成106DNA序列/cm2,合成长度60 nt(已商业化) | 与传统亚磷酸酰胺法单体试剂不兼容,光干扰影响偶联效率 |
| 光敏抗蚀层合成法 | 光通过掩模(或者虚拟掩模)照射特定的阵点。将光敏抗蚀层清除,采用常用的酸脱保护剂,将暴露区域5′-羟基保护基团脱除,被活化的5′端羟基可以进行偶联、封闭、氧化反应 | 光刻分辨率高,与传统亚磷酸酰胺法单体试剂兼容。可合成106DNA序列/cm2,合成长度20 nt | 涂胶、显影、去胶等复杂的光刻工艺,极易污染合成芯片,同时影响被保护寡核苷酸的稳定性 |
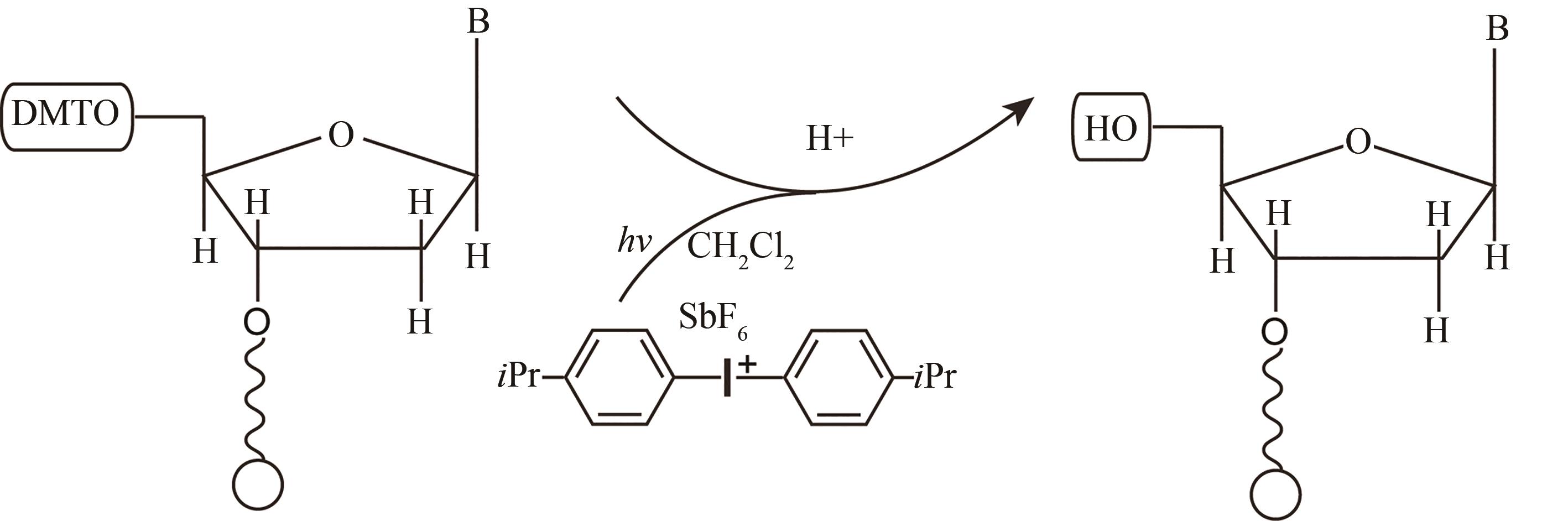
| 光致酸法 | 光通过掩模(或者虚拟掩模)照射特定的阵点,使阵点上物质发生光化学反应,产生的酸将5′端保护基团脱除,被活化的5′端羟基可以进行偶联、封闭、氧化反应 | 光刻分辨率高,单位面积上阵点数目多。可合成106DNA序列/cm2,合成长度120 nt | 与传统亚磷酸酰胺法脱保护试剂不兼容,需选择合适的光致酸试剂,需防止酸扩散污染 |
| 喷印合成法 | 在特定阵点上喷印单体进行偶联 | 与传统亚磷酸酰胺法兼容,合成效率高。可合成103DNA序列/cm2,合成长度120 nt | 喷头的机械定位精度要求高,喷印的位置无法做到完全重叠,不适合高密度DNA阵列合成 |
| 电致酸法 | 在特定阵点的电极通电,使阵点周围物质发生电化学反应,产生的酸将5′端保护基团脱除,被活化的5′端羟基可以进行偶联、封闭、氧化反应 | 半导体制造技术,合成区域定位准确。可合成105DNA序列/cm2,合成长度160 nt(已商业化) | 与传统亚磷酸酰胺法脱保护试剂不兼容,需选择合适的电致酸试剂,需防止酸扩散污染 |
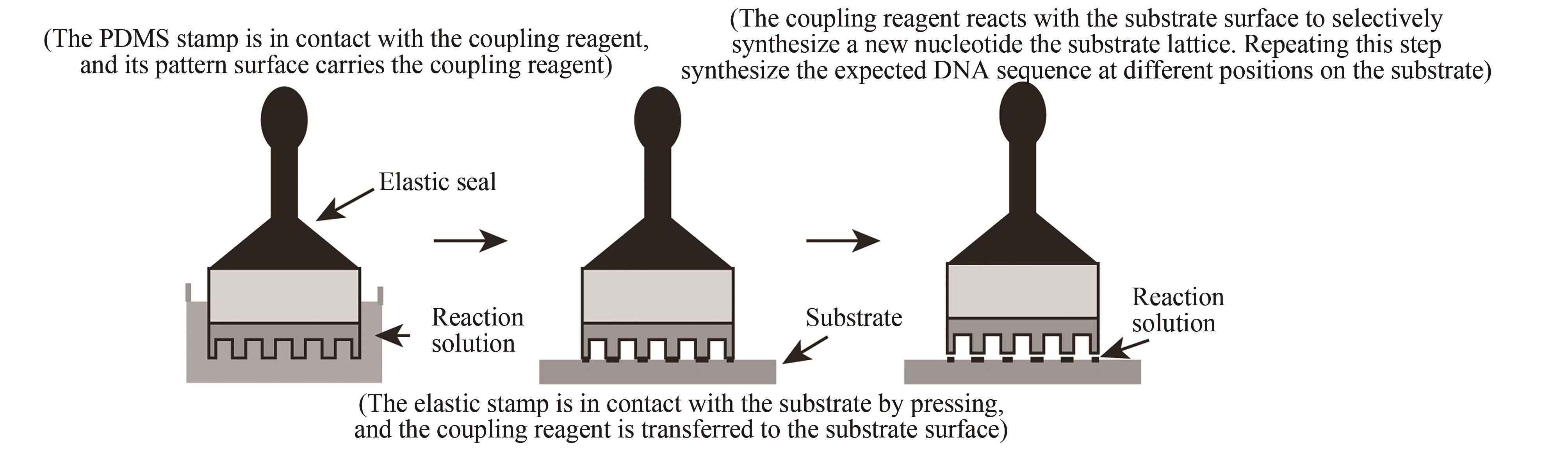
| 压印法 | 在特定阵点上压印单体进行偶联 | 与传统亚磷酸酰胺法兼容,合成效率高。可合成6×104DNA序列/cm2,合成长度60 nt(已商业化) | 压印机械定位精度要求高,两个接触面需高度平行 |
Tab. 1 Characteristics of in situ synthesis of microarray DNA
| 方法 | 技术特征 | 优点 | 缺点 |
|---|---|---|---|
| 原位光刻法 | 光通过掩模(或者虚拟掩模)照射特定的阵点,5'端保护基团发生光化学反应被直接脱除,被活化的5'-羟基可以进行偶联、封闭、氧化反应 | 光刻的分辨率高。可合成106DNA序列/cm2,合成长度60 nt(已商业化) | 与传统亚磷酸酰胺法单体试剂不兼容,光干扰影响偶联效率 |
| 光敏抗蚀层合成法 | 光通过掩模(或者虚拟掩模)照射特定的阵点。将光敏抗蚀层清除,采用常用的酸脱保护剂,将暴露区域5′-羟基保护基团脱除,被活化的5′端羟基可以进行偶联、封闭、氧化反应 | 光刻分辨率高,与传统亚磷酸酰胺法单体试剂兼容。可合成106DNA序列/cm2,合成长度20 nt | 涂胶、显影、去胶等复杂的光刻工艺,极易污染合成芯片,同时影响被保护寡核苷酸的稳定性 |
| 光致酸法 | 光通过掩模(或者虚拟掩模)照射特定的阵点,使阵点上物质发生光化学反应,产生的酸将5′端保护基团脱除,被活化的5′端羟基可以进行偶联、封闭、氧化反应 | 光刻分辨率高,单位面积上阵点数目多。可合成106DNA序列/cm2,合成长度120 nt | 与传统亚磷酸酰胺法脱保护试剂不兼容,需选择合适的光致酸试剂,需防止酸扩散污染 |
| 喷印合成法 | 在特定阵点上喷印单体进行偶联 | 与传统亚磷酸酰胺法兼容,合成效率高。可合成103DNA序列/cm2,合成长度120 nt | 喷头的机械定位精度要求高,喷印的位置无法做到完全重叠,不适合高密度DNA阵列合成 |
| 电致酸法 | 在特定阵点的电极通电,使阵点周围物质发生电化学反应,产生的酸将5′端保护基团脱除,被活化的5′端羟基可以进行偶联、封闭、氧化反应 | 半导体制造技术,合成区域定位准确。可合成105DNA序列/cm2,合成长度160 nt(已商业化) | 与传统亚磷酸酰胺法脱保护试剂不兼容,需选择合适的电致酸试剂,需防止酸扩散污染 |
| 压印法 | 在特定阵点上压印单体进行偶联 | 与传统亚磷酸酰胺法兼容,合成效率高。可合成6×104DNA序列/cm2,合成长度60 nt(已商业化) | 压印机械定位精度要求高,两个接触面需高度平行 |
| 1 | 王冬梅, 洪泂. 从碱基到人造生命——基因组的从头合成[J]. 生命的化学, 2011, 31(1): 13-20. |
| WANG D M, HONG J. From base to artificial life—chemical synthesis of genome[J]. Chemistry of Life, 2011, 31(1): 13-20. | |
| 2 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 3 | 李雷, 姜卫红, 覃重军,等. 合成生物学使能技术的研究进展[J]. 中国科学: 生命科学, 2015, 45(10): 950-968. |
| LI L, JIANG W H, QIN Z J, et al. Recent advances in the enabling technologies for synthetic biology[J]. SCIENTIA SINICA Vitae, 2015, 45(10): 950-968. | |
| 4 | 李诗渊, 赵国屏, 王金. 合成生物学技术的研究进展——DNA合成、组装与基因组编辑[J]. 生物工程学报, 2017, 33(3): 343-360. |
| LI S Y, ZHAO G P, WANG J. Enabling technologies in synthetic biology—DNA synthesis, assembly and editing[J]. Chinese Journal of Biotechnology, 2017, 33(3): 343-360. | |
| 5 | 陈婧, 肖鹏峰. 高通量DNA合成测序化学研究进展[J]. 生物过程, 2012, 2(1): 1-6. |
| CHEN J, XIAO P F. Advance in sequence chemistry of high-throughput DNA sequencing by synthesis[J]. Bioprocess, 2012, 2(1): 1-6. | |
| 6 | ABRAMOVA T. Frontiers and approaches to chemical synthesis of oligodeoxyribonucleotides[J]. Molecules, 2013, 18(1): 1063-1075. |
| 7 | CARUTHERS M H. The chemical synthesis of DNA/RNA: our gift to science[J]. The Journal of Biological Chemistry, 2013, 288(2): 1420-1427. |
| 8 | REESE C B. Oligo-and poly-nucleotides: 50 years of chemical synthesis[J]. Organic & Biomolecular Chemistry, 2005, 3(21): 3851-3868. |
| 9 | ABRAMOVA T V, SILNIKOV V N. Synthesis and properties of carbohydrate-phosphate backbone-modified oligonucleotide analogues and nucleic acid mimetics[J]. Russian Chemical Reviews, 2011, 80(5): 429-452. |
| 10 | WATTS J K, COREY D R. Silencing disease genes in the laboratory and the clinic[J]. The Journal of Pathology, 2012, 226(2): 365-379. |
| 11 | HUGHES R A, ELLINGTON A D. Synthetic DNA synthesis and assembly: putting the synthetic in synthetic biology[J]. Cold Spring Harbor Perspectives In Biology, 2017, 9(1):1-17. |
| 12 | MCDANIEL R, WEISS R. Advances in synthetic biology: on the path from prototypes to applications[J]. Current Opinion Biotechnology, 2005, 16(4): 476-483. |
| 13 | KOBAYASHI H, KAERN M, ARAKI M, et al. Programmable cells: interfacing natural and engineered gene networks[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(22): 8414-8419. |
| 14 | TIAN J, GONG H, SHENG N, et al. Accurate multiplex gene synthesis from programmable DNA microchips[J]. Nature, 2004, 432(7020): 1050-1054. |
| 15 | GIBSON D G, BENDERS G A, ANDREWS-PFANNKOCH C, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome[J]. Science, 2008, 319(5867): 1215-1220. |
| 16 | SMITH H O, HUTCHISON C A, PFANNKOCH C, et al. Generating a synthetic genome by whole genome assembly: PhiX174 bacteriophage from synthetic oligonucleotides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(26): 15440-15445. |
| 17 | CELLO J, PAUL A V, WIMMER E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template[J]. Science, 2002, 297(5583): 1016-1018. |
| 18 | CHAN L Y, KOSURI S, ENDY D. Refactoring bacteriophage T7[J]. Molecular Systems Biology, 2005, 1: 1-10. |
| 19 | 卢俊南, 罗周卿, 姜双英,等. DNA的合成、组装及转移技术[J]. 中国科学院院刊, 2018, 33(11):1174-1183. |
| LU J N, LUO Z Q, JIANG S Y, et al. Synthesis, assembly and transfer of DNA[J]. Bulletin of Chinese Academy of Sciences, 2018, 33 (11) :1174-1183. | |
| 20 | ZHRINOV V V, RASIC D. 2018 semiconductor synthetic biology roadmap[EB/OL]. [2021-05-24]. . |
| 21 | ERLICH Y, ZIELINSKI D, ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture[J]. Science, 2017, 355(6328): 950-954. |
| 22 | ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36(3): 242-248. |
| 23 | KIM J, BAE J H, BAYM M, et al. Metastable hybridization-based DNA information storage to allow rapid and permanent erasure[J]. Nature Communications, 2020, 11(1): 5008. |
| 24 | HAO M, QIAO H, GAO Y, et al. A mixed culture of bacterial cells enables an economic DNA storage on a large scale[J]. Communications Biology, 2020, 3(1): 416. |
| 25 | LEE H, WIEGAND D J, GRISWOLD K, et al. Photon-directed multiplexed enzymatic DNA synthesis for molecular digital data storage[J]. Nature Communications, 2020, 11(1): 5246. |
| 26 | CEZE L, NIVALA J, STRAUSS K. Molecular digital data storage using DNA[J]. Nature Reviews Genetics, 2019, 20(8): 456-466. |
| 27 | MEISER L C, ANTKOWIAK P L, KOCH J, et al. Reading and writing digital data in DNA[J]. Nature Protocols, 2020, 15(1): 86-101. |
| 28 | RUTTEN M G T A, VAANDRAGER F W, ELEMANS J A A W, et al. Encoding information into polymers[J]. Nature Reviews Chemistry, 2018, 2(11): 365-381. |
| 29 | CHEN Y J, TAKAHASHI C N, ORGANICK L, et al. Quantifying molecular bias in DNA data storage[J]. Nature Communications, 2020, 11(1): 3264. |
| 30 | HECKEL R, MIKUTIS G, GRASS R N. A characterization of the DNA data storage channel[J]. Scientific Reports, 2019, 9(1): 9663. |
| 31 | ANAVY L, VAKNIN I, ATAR O, et al. Data storage in DNA with fewer synthesis cycles using composite DNA letters[J]. Nature Biotechnology, 2019, 37(10): 1229-1236. |
| 32 | TABATABAEI S K, WANG B, ATHREYA N B M, et al. DNA punch cards for storing data on native DNA sequences via enzymatic nicking[J]. Nature Communications, 2020, 11(1): 1742. |
| 33 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications[J]. Nature Methods, 2014, 11(5): 499-507. |
| 34 | LIM H, CHO N, AHN J, et al. Highly selective retrieval of accurate DNA utilizing a pool of in situ-replicated DNA from multiple next-generation sequencing platforms[J]. Nucleic Acids Research, 2018, 46(7): e40. |
| 35 | KOSURI S, EROSHENKO N, LEPROUST E M, et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips[J]. Nature Biotechnology, 2010, 28(12): 1295-1299. |
| 36 | MATZAS M, STÄHLER P F, KEFER N, et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing[J]. Nature Biotechnology, 2010, 28(12): 1291-1294. |
| 37 | CLEARY M A, KILIAN K, WANG Y, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis[J]. Nature Methods, 2004, 1(3): 241-248. |
| 38 | LEE C C, SNYDER T M, QUAKE S R. A microfluidic oligonucleotide synthesizer[J]. Nucleic Acids Research, 2010. 38(8): 2514-2521. |
| 39 | MATTEUCCI M D, CARUTHERS M H. Synthesis of deoxyoligonucleotides on a polymer support[J]. Journal of the American Chemical Society, 1981, 103(11): 3185-3191. |
| 40 | TIAN J D, MA K S, SAAEM I. Advancing high-throughput gene synthesis technology[J]. Molecular BioSystems, 2009, 5(7): 714-722. |
| 41 | CARUTHERS M H, BEAUCAGE S L, BECKER C, et al. Deoxyoligonucleotide synthesis via the phosphoramidite method[J]. Gene Amplification and Analysis, 1983, 3: 1-26. |
| 42 | CARUTHERS M H, BARONE A D, BEAUCAGE S L, et al. Chemical synthesis of deoxyoligonucleotides by the phosphoramidite method[J]. Methods in Enzymology, 1987, 154: 287-313. |
| 43 | CARUTHERS M H. Gene synthesis machines: DNA chemistry and its uses[J]. Science, 1985, 230(4723): 281-285. |
| 44 | LIETARD J, SCHAUDY E, HOLZ K, et al. High-density DNA and RNA microarrays-photolithographic synthesis, hybridization and preparation of large nucleic acid libraries[J]. Journal of Visualized Experiments, 2019(150): 1-13. |
| 45 | MATTEUCCI M D, CARUTHERS M H. Synthesis of deoxyoligonucleotides on a polymer support[J]. Journal of the American Chemical Society, 1981, 103(11): 3185-3191. |
| 46 | EROSHENKO N, KOSURI S, MARBLESTONE A H, et al. Gene assembly from chip-synthesized oligonucleotides[J]. Current Protocols Chemical Biology, 2012, 4(1): 1-24. |
| 47 | 汤建新, 陈洪, 何农跃. 活版印刷技术原位合成DNA微阵列[J]. 中国科学(B辑化学), 2006, 36(6): 493-500. |
| TANG J X, CHEN H, HE N Y. In situ synthesis of DNA microarrays by letterpress printing[J]. SCIENCE IN CHINA Ser. B Chemistry, 2006, 36(6):493-500. | |
| 48 | MASKOS U, SOUTHERN E M. A novel method for the parallel analysis of multiple mutations in multiple samples[J]. Nucleic Acids Research, 1993, 21(9): 2269-2270. |
| 49 | SOUTHERN E M, MASKOS U, ELDER J K. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models[J]. Genomics, 1992, 13(4): 1008-1017. |
| 50 | SOUTHERN E M, CASE-GREEN S C, EIDER J K, et al. Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids[J]. Nucleic Acids Research, 1994, 22(8): 1368-1373. |
| 51 | GAO X, GULARI E, ZHOU X. In situ synthesis of oligonucleotide microarrays[J]. Biopolymers, 2004, 73(5): 579-596. |
| 52 | WANG C, TRAU D. A portable generic DNA bioassay system based on in situ oligonucleotide synthesis and hybridization detection[J]. Biosensors Bioelectronics, 2011, 26(5): 2436-2441. |
| 53 | SUMMERER D, HEVRONI D, JAIN A, et al. A flexible and fully integrated system for amplification, detection and genotyping of genomic DNA targets based on microfluidic oligonucleotide arrays[J]. Nature Biotechnology, 2010, 27(2): 149-155. |
| 54 | KIM C, KAYSEN J, RICHMOND K, et al. Progress in gene assembly from a MAS-driven DNA microarray[J]. Microelectronic Engineering, 2006, 83(4/5/6/7/8/9): 1613-1616. |
| 55 | RICHMOND K E, LI M H, RODESCH M J, et al. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis[J]. Nucleic Acids Research, 2004, 32(17): 5011-5018. |
| 56 | ZHOU X C, CAI S Y, HONG A L, et al. Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences[J]. Nucleic Acids Research, 2004, 32(18): 5409-5417. |
| 57 | FODOR S P, READ J L, PIRRUNG M C, et al. Light-directed, spatially addressable parallel chemical synthesis[J]. Science, 1991, 251(4995): 767-773. |
| 58 | LIPSHUTZ R J, FODOR S P A, GINGERAS T R, et al. High density synthetic oligonucleotide arrays[J]. Nature Genetics, 1999, 21: 20-24. |
| 59 | PEASE A C, SOLAS D, SULLIVAN E J, et al. Light-generated oligonucleotide arrays for rapid DNA sequence analysis[J]. Proceedings of the National Academy of Sciences of the United States of America, 1994, 91(11): 5022-5026. |
| 60 | FODOR S P, RAVA R P, HUANG X C, et al. Multiplexed biochemical assays with biological chips[J]. Nature, 1993, 364(6437): 555-556. |
| 61 | DALMA-WEISZHAUSZ D, WARRINGTON D, JANET T, et al. The affymetrix geneChip® platform: an overview[J]. Methods Enzymol, 2006, 410: 3-28. |
| 62 | SINGH-GASSON S, GREEN R D, YUE Y, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array[J]. Nature Biotechnology, 1999, 17(10): 974-978. |
| 63 | CERRINA F, BLATTNER F, HUANG W, et al. Biological lithography: development of a maskless microarray synthesizer for DNA chips[J]. Microelectronic Engineering, 2002, 61/62: 33-40. |
| 64 | NUWAYSIR E F, HUANG W, ALBERT T J, et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography[J]. Genome Research, 2002, 12(11): 1749-1755. |
| 65 | AGBAVWE C, KIM C, HONG D, et al. Efficiency, error and yield in light-directed maskless synthesis of DNA microarrays[J]. Journal of Nanobiotechnology, 2011, 9: 57. |
| 66 | SACK M, KRETSCHY N, ROHM B, et al. Simultaneous light-directed synthesis of mirror-image microarrays in a photochemical reaction cell with flare suppression[J]. Analytical Chemistry, 2013, 85(18): 8513-8517. |
| 67 | SACK M, HÖLZ K, HOLIK A K, et al. Express photolithographic DNA microarray synthesis with optimized chemistry and high-efficiency photolabile groups[J]. Journal of Nanobiotechnology, 2016, 14(1): 1-13. |
| 68 | KRETSCHY N, HOLIK A K, SOMOZA V, et al. Next-generation o-nitrobenzyl photolabile groups for light-directed chemistry and microarray synthesis[J]. Angewandte Chemie International Edition, 2015, 54(29): 8555-8559. |
| 69 | LACKEY J G, MITRA D, SOMOZA M M, et al. Acetal levulinyl ester (ALE) groups for 2'-hydroxyl protection of ribonucleosides in the synthesis of oligoribonucleotides on glass and microarrays[J]. Journal of the American Chemical Society, 2009, 131(24): 8496-8502. |
| 70 | MCGALL G, LABADIE J, BROCK P, et al. Light-directed synthesis of high-density oligonucleotide arrays using semiconductor photoresists[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(24): 13555-13560. |
| 71 | KIM C, LI M, RODESCH M, et al. Biological lithography: improvements in DNA synthesis methods[J]. Journal of Vacuum Science & Technology B, 2004, 22(6): 3163-3167. |
| 72 | PAWLOSKI A R, MCGALL G, KUIMELIS R G, et al. Photolithographic synthesis of high-density DNA probe arrays: challenges and opportunities[J]. Journal of Vacuum Science & Technology B, 2007, 25(6): 2537-2546. |
| 73 | PEASE A C, SOLAS D, SULLIVAN E J, et al. Light-generated oligonucleotide arrays for rapid DNA-sequence analysis[J]. Proceedings of the National Academy of Sciences of the United States of America, 1994, 91(11): 5022-5026. |
| 74 | ALBERT T J, NORTON J, OTT M, et al. Light-directed 5'→3' synthesis of complex oligonucleotide microarrays[J]. Nucleic Acids Research, 2003, 31(7): e35. |
| 75 | GAO X L, LEPROUST E, ZHANG H, et al. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids[J]. Nucleic Acids Research, 2001, 29(22): 4744-4750. |
| 76 | SRIVANNAVIT O, GULARI M, HUA Z, et al. Microfluidic reactor array device for massively parallel in situ synthesis of oligonucleotides[J]. Sensors and Actuators B: Chemical, 2009, 140(2): 473-481. |
| 77 | LEPROUST E, PELLOIS J P, YU P L, et al. Digital light-directed synthesis. A microarray platform that permits rapid reaction optimization on a combinatorial basis[J]. Journal of Combinatorial Chemistry, 2000, 2(4): 349-354. |
| 78 | KOMOLPIS K, SRIVANNAVIT O, GULARI E. Light-directed simultaneous synthesis of oligopeptides on microarray substrate using a photogenerated acid[J]. Biotechnology Progress, 2002, 18(3): 641-646. |
| 79 | HUGHES T R, MAO M, JONES A R, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer[J]. Nature Biotechnology, 2001, 19(4): 342-347. |
| 80 | YANG Y H, DUDOIT S, LUU P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation[J]. Nucleic Acids Research, 2002, 30(4): e15. |
| 81 | BAUM M, BIELAU S, RITTNER N, et al. Validation of a novel, fully integrated and flexible microarray benchtop facility for gene expression profiling[J]. Nucleic Acids Research, 2003, 31(23): e151. |
| 82 | HUGHES T R, MAO M, JONES A R, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer[J]. Nature Biotechnology, 2001, 19(4): 342-347. |
| 83 | MRKSICH M, WHITESIDES G M. Patterning self-assembled monolayers using microcontact printin-a new technology for biosensors[J]. Trends in Biotechnology, 1995, 13(6): 228-235. |
| 84 | MRKSICH M, CHEN C S, XIA Y, et al. Controlling cell attachment on contoured surfaces with self-assembled monolayers of alkanethiolates on gold[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(20): 10775-10778. |
| 85 | JAMES C D, DAVIS R C, KAM L, et al. Patterned protein layers on solid substrates by thin stamp microcontact printing[J]. Langmuir, 1998, 14(4): 741-744. |
| 86 | MRKSICH M, DIKE L E, TIEN J, et al. Using microcontact printing to pattern the attachment of mammalian cells to self-assembled monolayers of alkanethiolates on transparent films of gold and silver[J]. Experimental Cell Research, 1997, 235(2): 305-313. |
| 87 | YAN L, ZHAO X M, WHITESIDES G M. Patterning a preformed, reactive SAM using microcontact printing[J]. Journal of the American Chemical Society, 1998, 120(24): 6179-6180. |
| 88 | YAN L, HUCK W T S, ZHAO X M, et al. Patterning thin films of poly(ethylene imine) on a reactive SAM using microcontact printing[J]. Langmuir, 1999, 15(4): 1208-1214. |
| 89 | LAHIRI J, OSTUNI E, WHITESIDES G M. Patterning ligands on reactive SAMs by microcontact printing[J]. Langmuir, 1999, 15(6): 2055-2060. |
| 90 | 陆祖宏, 何农跃, 赵雨杰,等. 制备化合物微阵列芯片的方法及由该方法制备的化合物微阵列芯片: CN1274758[P]. 2004-07-28. |
| LU Z H, HE N Y, ZHAO Y J, et al. Method for preparing compound microarray chip and compound microarray chip prepared by the method: CN1274758[P]. 2004-07-28. | |
| 91 | 肖鹏峰, 何农跃, 贺全国,等. 分子印章法DNA芯片的合成[J]. 中国科学(B辑化学), 2001, 31(5): 399-404. |
| XIAO P F, HE N Y, HE Q G, et al. Synthesis of DNA chip by molecular seal method[J]. SCIENCE IN CHINA Ser. B Chemistry, 2001, 31(5): 399-404. | |
| 92 | 肖鹏峰, 陆祖宏, 刘正春,等. 分子印章法DNA芯片合成(Ⅲ)——反应条件的优化[J]. 科学通报, 2002, 47(3): 189-192. |
| XIAO P F, LU Z H, LIU Z C, et al. DNA chip synthesis by molecular seal method (Ⅲ)-optimization of reaction conditions[J]. Chinese Science Bulletin, 2002, 47(3): 189-192. | |
| 93 | 肖鹏峰, 何农跃, 朱纪军,等. 分子印章法DNA芯片的合成(Ⅰ)[J]. 东南大学学报(自然科学版), 2001, 31(2): 756-762. |
| XIAO P F, HE N Y, ZHU J J, et al. DNA microarray synthesis using PDMS molecular stamps (Ⅰ): the fixation and shrinkage of PDMS molecular stamps[J]. Journal of Southeast University (Natural Science Edition), 2001, 31(2): 756-762. | |
| 94 | XIA Y, WHITESIDES G M. Soft lithography[J]. Annual Review of Materials Science, 1998, 28(1): 153-184. |
| 95 | 孙啸, 王晔, 赵雨杰,等. 一种高密度基因芯片设计的新方法[J]. 电子学报, 2001, 29(3): 293-296. |
| SUN X, WANG Y, ZHAO Y J, et al. A new method of high-density gene chip design[J]. Acta Electronica Sinica, 2001, 29(3): 293-296. | |
| 96 | 王轶文, 徐吉庆, 陈扬,等. 电解致酸脱保护DNA在片合成研究[J]. 东南大学学报(自然科学版), 2002, 32(1): 77-80. |
| WANG Y W, XU J Q, CHEN Y, et al. Study on on-chip synthesis of electrolytic acid-deprotected DNA[J]. Journal of Southeast University (Natural Science Edition), 2002, 32(1): 77-80. | |
| 97 | EGELAND R D, SOUTHERN E M. Electrochemically directed synthesis of oligonucleotides for DNA microarray fabrication[J]. Nucleic Acids Research, 2005, 33(14): e125. |
| 98 | MAURER K, YAZVENKO N, WILMOTH J, et al. Use of a multiplexed CMOS microarray to optimize and compare oligonucleotide binding to DNA probes synthesized or immobilized on individual electrodes[J]. Sensors, 2010, 10(8): 7371-7385. |
| 99 | GHINDILIS A L, SMITH M W, SCHWARZKOPF K R, et al. CombiMatrix oligonucleotide arrays: genotyping and gene expression assays employing electrochemical detection[J]. Biosensors & Bioelectronics, 2007, 22(9/10): 1853-1860. |
| 100 | COOPER J, YAZVENKO N, PEYVAN K, et al. Targeted deposition of antibodies on a multiplex CMOS microarray and optimization of a sensitive immunoassay using electrochemical detection[J]. PLoS One, 2010, 5(3): e9781. |
| 101 | MAURER K, COOPER J, CARABALLO M, et al. Electrochemically generated acid and its containment to 100 micron reaction areas for the production of DNA microarrays[J]. PLoS One, 2006, 1: e34. |
| 102 | CHOW B Y, EMIG C J, JACOBSON J M. Photoelectrochemical synthesis of DNA microarrays[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(36): 15219-15224. |
| 103 | CHOI Y, RYU T, LEE A C, et al. High information capacity DNA-based data storage with augmented encoding characters using degenerate bases[J]. Scientific Reports, 2019, 9(1): 6582. |
| 104 | GUO Y, DONG J, ZHOU T, et al. YeastFab: the design and construction of standard biological parts for metabolic engineering in Saccharomyces cerevisiae [J]. Nucleic Acids Research, 2015, 43(13): e88. |
| 105 | QIN Y R, TAN C, LIN J W, et al. EcoExpress—highly efficient construction and expression of multicomponent protein complexes in escherichia coli[J]. ACS Synthetic Biology, 2016, 5(11): 1239-1246. |
| 106 | PALLUK S, ARLOW D H, ROND T D, et al. De novo DNA synthesis using polymerase-nucleotide conjugates[J]. Nature Biotechnology, 2018, 36(7): 645-650. |
| 107 | JENSEN M A, DAVIS R W. Template-independent enzymatic oligonucleotide synthesis (TiEOS): its history, prospects, and challenges[J]. Biochemistry, 2018, 57(12): 1821-1832. |
| 108 | SINDELAR L E, JAKLEVIC J M. High-throughput DNA synthesis in a multichannel format[J]. Nucleic Acids Research, 1995, 23(6): 982-987. |
| 109 | LASHKARI D A, HUNICKESMITH S P, NORGREN R M, et al. An automated multiplex oligonucleotide synthesizer-development of high-throughput, low-cost DNA-synthesis[J]. Proceedings of the National Academy of Sciences of the United States of America, 1995, 92(17): 7912-7915. |
| 110 | CHENG J Y, CHEN H H, KAO Y S, et al. High throughput parallel synthesis of oligonucleotides with 1536 channel synthesizer[J]. Nucleic Acids Research, 2002, 30(18): e93. |
| 111 | CHENG J Y, CHEN H Y. Microfluidic ARray Synthesizer (MArS) for rapid preparation and hybridization of custom DNA microarray[J]. Biotechnology and Bioengineering, 2009, 104(2): 400-407. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [7] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [8] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [9] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [10] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [11] | ZHANG Xuanliang, LI Qingting, WANG Fei. Data writing in DNA storage systems [J]. Synthetic Biology Journal, 2024, 5(5): 1125-1141. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||