Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (3): 412-427.DOI: 10.12211/2096-8280.2020-083
Previous Articles Next Articles
Chamaeleo: an integrated evaluation platform for DNA storage
PING Zhi1,2,3, ZHANG Haoling1,2,3, CHEN Shihong4,5, NI Ming1,3, XU Xun1,2,3, ZHU Sha6,7, SHEN Yue1,2,3,5
- 1.BGI-Shenzhen,Shenzhen 518083,Guangdong,China
2.Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
3.Guangdong Provincial Key Laboratory of Genome Read and Write,BGI-Shenzhen,Shenzhen 518120,Guangdong,China
4.Guangdong Provincial Academician Workstation of BGI Synthetic Genomics,BGI-Shenzhen,Shenzhen 518120,Guangdong,China
5.China National GeneBank,BGI-Shenzhen,Shenzhen 518120,Guangdong,China
6.Big Data Institute,University of Oxford,Oxford,OX3 7LF,United Kingdom
7.TAICHI AI Ltd. ,London,N1 7GU,United Kingdom
-
Received:2020-12-01Revised:2020-12-31Online:2021-07-13Published:2021-06-30 -
Contact:ZHU Sha, SHEN Yue
Chamaeleo: DNA存储碱基编解码算法的可拓展集成与系统评估平台
平质1,2,3, 张颢龄1,2,3, 陈世宏4,5, 倪鸣1,3, 徐讯1,2,3, 朱砂6,7, 沈玥1,2,3,5
- 1.深圳华大生命科学研究院,广东 深圳 518083
2.深圳市合成生物学创新研究院,中国科学院深圳先进技术研究院,广东 深圳 518055
3.广东省高通量基因组测序与合成编辑应用重点实验室,深圳华大生命科学研究院,广东 深圳 518120
4.广东省 )华大基因合成基因组学院士工作站,深圳华大基因科技有限公司,广东 深圳 518120
5.深圳国家基因库,广东 深圳 518120
6.英国牛津大学大数据研究所,牛津 OX3 7LF
7.英国TAICHI AI Ltd. ,伦敦 N1 7GU
-
通讯作者:朱砂,沈玥 -
作者简介:平质 (1987—),男,博士,助理研究员。研究方向为合成生物学、DNA存储、生物信息分析算法。 E-mail:pingzhi@genomics.cn张颢龄 (1996—),男,助理研究员。研究方向为合成生物学、计算机科学、神经网络。 E-mail:zhanghaoling@genomics.com朱砂 (1985—),男,博士,罗氏(英国)资深统计分析师。研究方向为生物遗传模型相关的概率论。长期从事计算机统计方法研究和程序开发。E-mail:sha.joe.zhu@gmail.com沈玥 (1986—),女,博士,研究员。研究方向为合成生物学、合成基因组学、DNA合成技术与工具开发。E-mail:shenyue@genomics.cn -
基金资助:国家重点研发计划(2020YFA0712100);广东省高通量基因组测序与合成编辑应用重点实验室项目(2017B030301011);广东省华大基因合成基因组学院士工作站项目(2017B090904014)
CLC Number:
Cite this article
PING Zhi, ZHANG Haoling, CHEN Shihong, NI Ming, XU Xun, ZHU Sha, SHEN Yue. Chamaeleo: an integrated evaluation platform for DNA storage[J]. Synthetic Biology Journal, 2021, 2(3): 412-427.
平质, 张颢龄, 陈世宏, 倪鸣, 徐讯, 朱砂, 沈玥. Chamaeleo: DNA存储碱基编解码算法的可拓展集成与系统评估平台[J]. 合成生物学, 2021, 2(3): 412-427.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-083
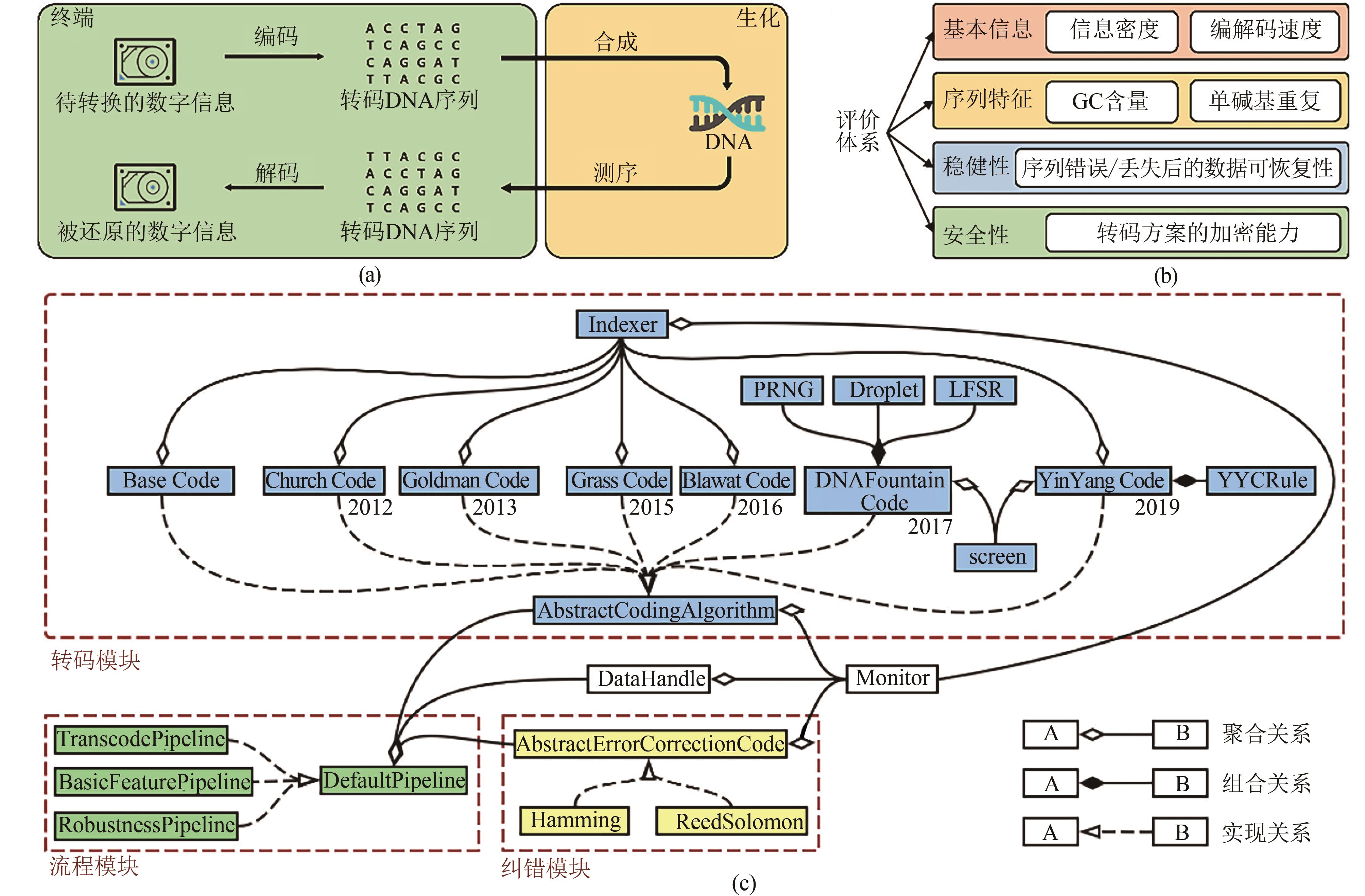
Fig. 1 Brief introduction of Chamaeleo[(a) General procedure of DNA storage includes in-silico transcoding and experimental DNA synthesis and sequencing. Chamaeleo focuses on in-silico transcoding part and connect the related technologies. (b) The evaluation system created by Chamaeleo quantitatively analyzes coding schemes from multiple aspects. (c) Software architecture and relations between modules and classes. The source code is available at https://github.com/ntpz870817/Chamaeleo, which can be also installed by the command of pip.exe, "pip install chamaeleo"]

Fig. 2 Structure of output DNA sequences and their net information densities[(a) Basic design of output DNA sequences with/without optional error-correction codes (Hamming code and RS code). (b) Distribution of net information density using different coding schemes. The setting of parameters is identical to that of original report: For Goldman's coding scheme, the Huffman tree used in the evaluation process is set as the same in Goldman, et al[4]. For DNA Fountain scheme, the degree distribution tuning parameter (δ = 0.05 and c_dist = 0.1) and redundancy (7%) used in the evaluation process is set as the same in Erlich, et al[6]. For Yin-Yang coding scheme, rule No. 888 was used as reported in Ping, et al[7]]
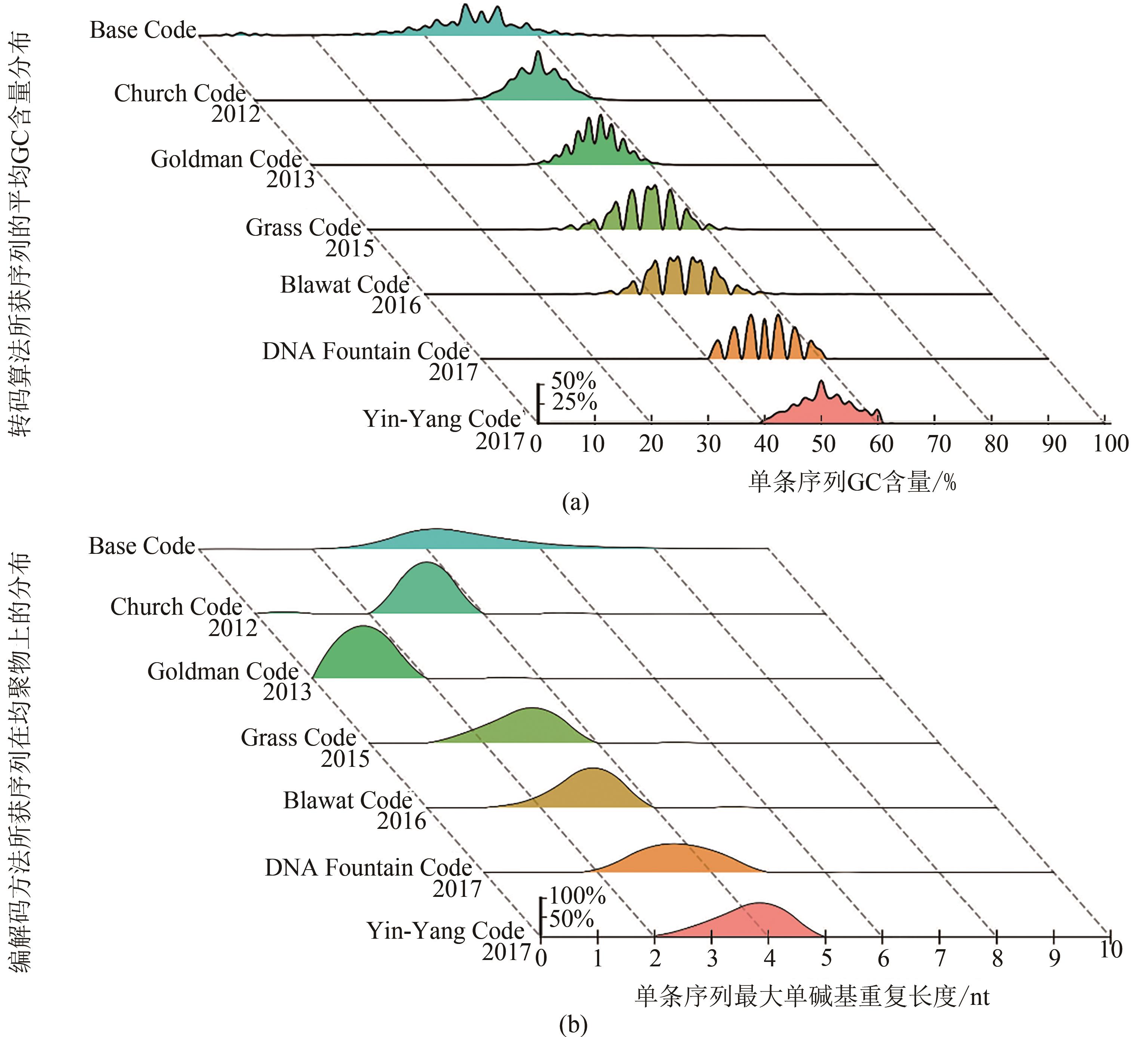
Fig. 3 Compatibility evaluation of coding schemes[Distribution of GC content (a) and maximum homopolymer length (b) of DNA sequences from transcoding of 10 test-files by different coding schemes. For Base coding in (b), the maximum homopolymer length is 42 and not shown in this panel for a clarity purpose]

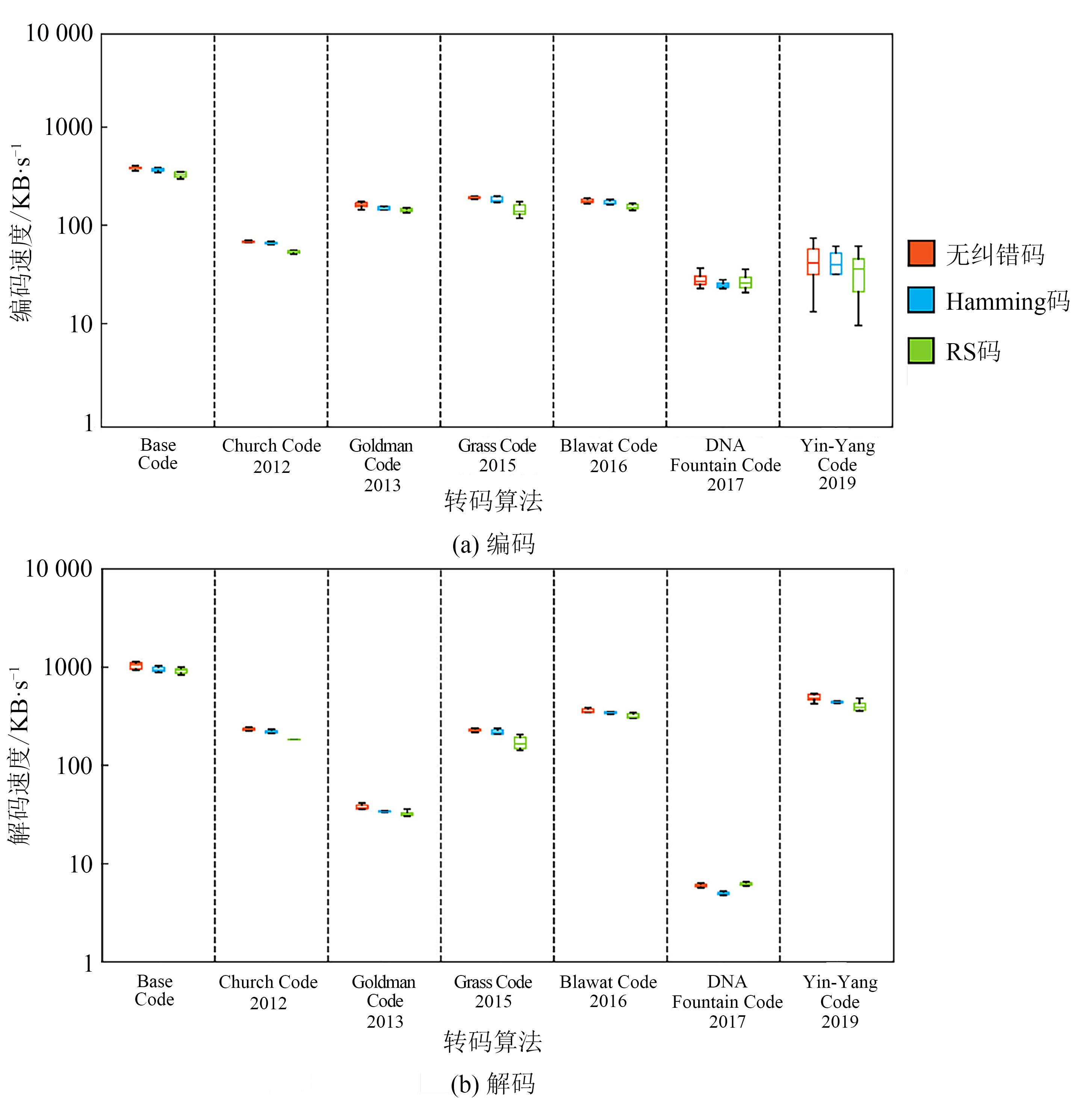
Fig. 4 Robustness evaluation of coding schemes[Distribution of file retrieval rate under condition of (a) 1% random sequence loss and (b) 3% of nucleotide error (1% for insertion/deletion/substitution, respectively). Figures on the right is the zoom in part for a closer view]

Fig. 5 Visualization of different coding schemes using Graph-theory-based presentation[Red dot represents starting point or virtual starting point (for Goldman & Yin-Yang coding schemes). During each step of encoding, with known previous base (node) and input bit value (arrow), the current base would be obtained from graph. During each step of decoding, with known previous and current nodes (base), the bit value (arrow) would be obtained from graph]
coding_scheme = Church(need_logs=True) error_correction = ReedSolomon(check_size=3, need_logs=True) pipeline = TranscodePipeline(coding_scheme=coding_scheme, error_correction=error_correction, need_logs=True) pipeline.transcode(direction="t_c", input_path="s.txt", output_path="t.dna", segment_length=120, index=True) pipeline.transcode(direction="t_s", input_path="t.dna", output_path="t.txt", index=True) pipeline.output_records(type="string") |
Tab.S1 Example commands to invoke transcoding process
coding_scheme = Church(need_logs=True) error_correction = ReedSolomon(check_size=3, need_logs=True) pipeline = TranscodePipeline(coding_scheme=coding_scheme, error_correction=error_correction, need_logs=True) pipeline.transcode(direction="t_c", input_path="s.txt", output_path="t.dna", segment_length=120, index=True) pipeline.transcode(direction="t_s", input_path="t.dna", output_path="t.txt", index=True) pipeline.output_records(type="string") |
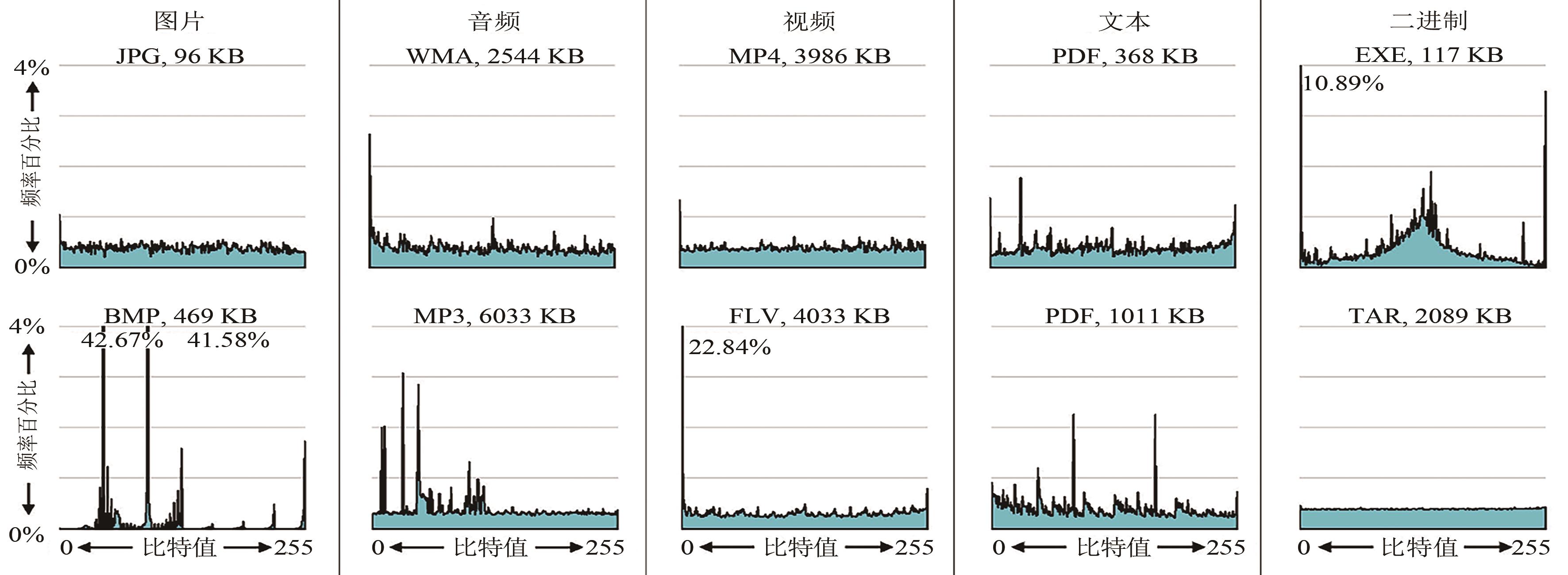
Fig. S1 Sizes and byte-frequency distributions of 10 typical test files[The 4 lines in each distribution indicates 1% to 4% from bottom to top. The probabilities of occurrence of few specific bytes exceed 4%, which are labeled with digits. The highest probability of occurrence is 42.67% ("00101110" in BMP file)]
| 1 | PING Z, MA D Z, HUANG X L, et al. Carbon-based archiving: current progress and future prospects of DNA-based data storage [J]. Gigascience, 2019, 8(6): gizo75. |
| 2 | DONG Y M, SUN F J, PING Z, et al. DNA storage: research landscape and future prospects [J]. National Science Review, 2020, 7(6): 1092-1107. |
| 3 | CHURCH G M, GAO Y, KOSURI S, Next-generation digital information storage in DNA [J]. Science, 2012, 337(6102): 1628. |
| 4 | GOLDMAN N, BERTONE P, CHEN S, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA [J]. Nature, 2013, 494(7435): 77-80. |
| 5 | GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes [J]. Angewandte Chemie International Edition, 2015, 54(8): 2552-2555. |
| 6 | ERLICH Y, ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture [J]. Science, 2017, 355(6328): 950-954. |
| 7 | PING Z, CHEN S, HUANG X, et al. Towards practical and robust DNA-based data archiving by codec system named 'Yin-Yang' [EB/OL]. [2021-05-26]. . |
| 8 | HAO M, QIAO H, GAO Y, et al. A mixed culture of bacterial cells enables an economic DNA storage on a large scale [J]. Communications Biology, 2020, 3(1): 416. |
| 9 | PRESS W H, HAWKINS J A, JONES S K, et al. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints [J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(31): 18489. |
| 10 | WANG Y X, NOOR-A-RAHIM M, GUNAWAN E, et al. Construction of bio-constrained code for DNA data storage [J]. IEEE Communications Letters, 2019, 23(6): 963-966. |
| 11 | NGUYEN T T, CAI K, IMMINK K A S, et al. Constrained coding with error control for DNA-based data storage [C]//2020 IEEE International Symposium on Information Theory (ISIT). 2020. |
| 12 | BLAWAT M, GAEDKE K, HUETTER I, et al. Forward error correction for DNA data storage [J]. Procedia Computer Science, 2016, 80: 1011-1022. |
| 13 | FOWLER M. Refactoring: improving the design of existing code [M]. Boston: Addison-Wesley Professional, 2018. |
| 14 | COX B J. Object-oriented programming: an evolutionary approach [M]. Boston: Addison-Wesley, 1986. |
| 15 | KOCH J, GANTENBEIN S, MASANIA K, et al. A DNA-of-things storage architecture to create materials with embedded memory [J]. Nature Biotechnology, 2020, 38(1): 39-43. |
| 16 | TANENBAUM A S BOS H. Modern operating systems [M]. London: Pearson, 2015. |
| 17 | SAYOOD K. Introduction to data compression [M]. Burlington: Morgan Kaufmann, 2017. |
| 18 | FENG L, FOH C H, JIANFEI C, et al. LT codes decoding: design and analysis [C]//2009 IEEE International Symposium on Information Theory. 2009. |
| 19 | YAZDI S H T, GABRYS R, MILENKOVIC O. Portable and error-free DNA-based data storage [J]. Scientific Reports, 2017, 7(1): 1-6. |
| 20 | ORGANICK L, CHEN Y J, ANG S D, et al. Probing the physical limits of reliable DNA data retrieval [J]. Nature Communications, 2020, 11(1): 1-7. |
| 21 | HECKEL R, MIKUTIS G, GRASS R N. A characterization of the DNA data storage channel [J]. Scientific Reports, 2019, 9(1): 9663. |
| 22 | MACKAY D J MAC KAY D J. Information theory, inference and learning algorithms [M]. Cambridge: Cambridge University Press, 2003. |
| 23 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications [J]. Nature Methods, 2014, 11(5): 499-507. |
| 24 | KULSKI J K. Next generation sequencing-advances, applications and challenges[M]. London: Intech Open, 2016: 3-60. |
| 25 | CHEN Y J, TAKAHASHI C N, ORGANICK L, et al. Quantifying molecular bias in DNA data storage [J]. Nature Communications, 2020, 11(1). DOI:http://doi.org/10.1038/s41467-020-16958-3 . |
| 26 | MOON T K. Error correction coding: mathematical methods and algorithms [M]. Hoboken: John Wiley & Sons, 2005. |
| 27 | STINSON D R, PATERSON M. Cryptography: theory and practice[M]. Boca Raton: CRC Press, 2018. |
| 28 | PASCHKE J, BURKERT J, FEHRIBACH R. Computing and estimating the number of n-ary Huffman sequences of a specified length [J]. Discrete Mathematics, 2011, 311(1): 1-7. |
| 29 | KOSHY T. Catalan numbers with applications [M]. Oxford: Oxford University Press, 2008. |
| 30 | AVAL J C. Multivariate fuss-catalan numbers [J]. Discrete Mathematics, 2008, 308(20): 4660-4669. |
| 31 | WEST D B. Introduction to graph theory [M]. Hoboken : Prentice Hall, 1996. |
| 32 | COMPEAU P E, PEVZNER P A, TESLER G. How to apply de Bruijn graphs to genome assembly [J]. Nature Biotechnology, 2011, 29(11): 987-991. |
| 33 | BOUCHET A. Greedy algorithm and symmetric matroids [J]. Mathematical Programming, 1987, 38(2): 147-159. |
| 34 | MILO R, SHEN-ORR S, ITZKOVITZ S, et al. Network motifs: simple building blocks of complex networks [J]. Science, 2002, 298(5594): 824-827. |
| 35 | BORNHOLT J, LOPEZ R, CARMEAN D M, et al. A DNA-based archival storage system [C]//Proceedings of the Twenty-First International Conference on Architectural Support for Programming Languages and Operating Systems. 2016. |
| 36 | CAFFERTY B J, TEN A S, FINK M J, et al. Storage of information using small organic molecules [J]. ACS Central Science, 2019, 5(5): 911-916. |
| 37 | KENNEDY E, ARCADIA C E, GEISER J, et al. Encoding information in synthetic metabolomes [J]. PLoS One, 2019, 14(7): e0217364. |
| [1] | Naicai ZHONG, Yuan CHEN, Wenfeng PAN, Xiaofeng SU, Jingwen LIAO, Jinyi ZHONG. Application progress of plasma microbial breeding technology in biofabrication [J]. Synthetic Biology Journal, 2025, (): 1-17. |
| [2] | Shuhan HUANG, He MA, Yunzi LUO. Research progress on biosynthesis of salidroside [J]. Synthetic Biology Journal, 2025, (): 1-16. |
| [3] | REN Jiawei, ZHANG Jinpeng, XU Guoqiang, ZHANG Xiaomei, XU Zhenghong, ZHANG Xiaojuan. Effect of terminators on the downstream transcript unit with gene expression in Escherichiacoli [J]. Synthetic Biology Journal, 2025, 6(1): 213-227. |
| [4] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [5] | Yongzhu LI, Yu CHEN. Advances and Prospects in Genome-Scale Models of Yeast [J]. Synthetic Biology Journal, 2025, (): 1-18. |
| [6] | LI Yifei, CHEN Ai, SUN Junsong, ZHANG Yi-Heng P. Job. Studies on hydrogenases for hydrogen production using in vitro synthetic enzymatic biosystems [J]. Synthetic Biology Journal, 2024, 5(6): 1461-1484. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | LIU Kuanqing, ZHANG Yi-Heng P.Job. Biological degradation and utilization of lignin [J]. Synthetic Biology Journal, 2024, 5(6): 1264-1278. |
| [9] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [10] | Xinyi GUO, Shuqi GUO, Shuwei LI, Ziyue JIAO, Qiang FEI. Progress in Biological Entity-Material Hybrid System for Low-Carbon Biosynthesis [J]. Synthetic Biology Journal, 2024, (): 1-14. |
| [11] | CHEN Guo-Qiang, TAN Dan. Reprogramming microbial chassis for low-cost bioprodcution of tailor-made polyhydroxyalkanoates [J]. Synthetic Biology Journal, 2024, 5(5): 1211-1226. |
| [12] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [13] | CHENG Xiaolei, LIU Tiangang, TAO Hui. Recent research progress in non-canonical biosynthesis of terpenoids [J]. Synthetic Biology Journal, 2024, 5(5): 1050-1071. |
| [14] | Jinchang LU, Yaokang WU, Xueqin LV, Long LIU, Jian CHEN, Yanfeng LIU. Green biomanufacturing of ceramide sphingolipids [J]. Synthetic Biology Journal, 2024, (): 1-23. |
| [15] | HAN Yizhao, GUO Jia, SHAO Yue. Stem cell-based synthetic development: cellular components, embryonic models, and engineering approaches [J]. Synthetic Biology Journal, 2024, 5(4): 734-753. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||