Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (3): 585-602.DOI: 10.12211/2096-8280.2024-084
• Invited Review • Previous Articles Next Articles
Advances and prospects in genome-scale models of yeast
LI Yongzhu1,2, CHEN Yu1
- 1.State Key Laboratory of Quantitative Synthetic Biology,Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2024-12-02Revised:2025-02-20Online:2025-06-27Published:2025-06-30 -
Contact:CHEN Yu
酵母基因组规模模型进展及发展趋势
李永珠1,2, 陈禹1
- 1.中国科学院深圳先进技术研究院,定量合成生物学全国重点实验室,深圳合成生物学创新研究院,广东 深圳 518055
2.中国科学院大学,北京 100049
-
通讯作者:陈禹 -
作者简介:李永珠 (2001—),女,硕士研究生。研究方向为代谢网络建模。E-mail:yz.li4@siat.ac.cn陈禹 (1990—),男,研究员,博士生导师。研究方向为系统生物学和定量合成生物学,致力于结合代谢网络建模和生物数据分析加速合成生物系统理性设计。E-mail:y.chen3@siat.ac.cn -
基金资助:国家重点研发计划(2023YFA0913900)
CLC Number:
Cite this article
LI Yongzhu, CHEN Yu. Advances and prospects in genome-scale models of yeast[J]. Synthetic Biology Journal, 2025, 6(3): 585-602.
李永珠, 陈禹. 酵母基因组规模模型进展及发展趋势[J]. 合成生物学, 2025, 6(3): 585-602.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-084
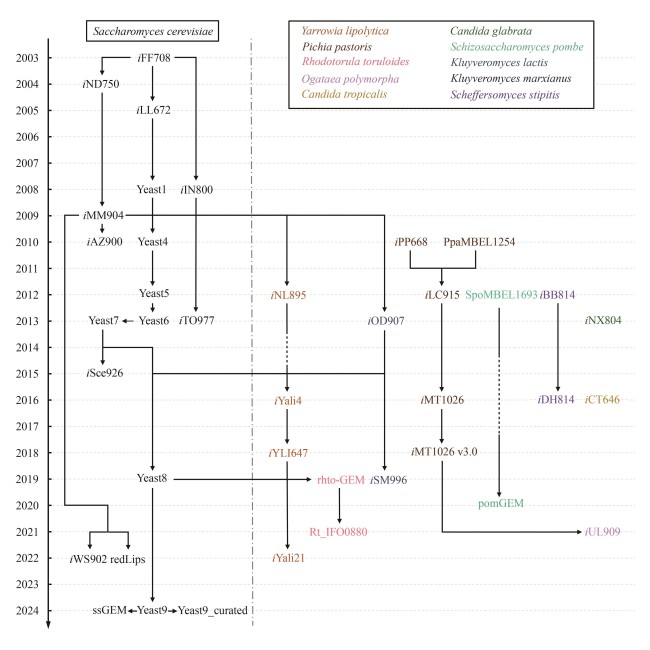
Fig. 1 Development of yeast GEMs[The figure shows the development of GEMs from 2003 to 2024 for S. cerevisiae (black) and other yeasts (colored). The arrows and lines indicate the relationships between different GEMs, where solid lines indicate direct inheritance, and dashed lines signify omissions of intermediate models between connections.]
| 模型版本 | 反应数 | 代谢物数 | 基因数 | 主要优化点 | 存在的问题 | 年份 | 参考文献 |
|---|---|---|---|---|---|---|---|
| Yeast1 | 1857 | 1168 | 832 | 第一代共识模型,统一代谢物注释 | 网络缺乏完整性、连通性,代谢反应覆盖率低 | 2008 | [ |
| Yeast4 | 2030 | 1481 | 924 | 增加脂质代谢反应,提高网络连通性 | 代谢反应覆盖率低 | 2010 | [ |
| Yeast5 | 2110 | 1655 | 918 | 增加鞘脂代谢反应,更新GPR关系 | 存在一定“阻塞反应”,引入假反应连通网络,不符合实际 | 2012 | [ |
| Yeast6 | 1888 | 1458 | 900 | 移除无明确功能或无参考来源的代谢物 | 模拟无氧生长存在问题,仍有一定“阻塞反应” | 2013 | [ |
| Yeast7 | 3493 | 2218 | 916 | 修正脂肪酸、甘油酯和甘油磷脂代谢,显著减少“阻塞反应”占比 | 基因数过少,无法充分整合多组学数据 | 2013 | [ |
| Yeast8 | 3949 | 2680 | 1133 | 对基因和反应进行大规模扩展,可引入蛋白质结构数据 | 反应质量和电荷不平衡,同工酶注释冗余,缺乏热力学约束 | 2019 | [ |
| Yeast9 | 4130 | 2805 | 1162 | 新增标准吉布斯自由能数据,提升反应准确性 | 无法精确表现细胞器代谢活动,多组学综合分析准确率较低 | 2024 | [ |
Table 1 Summary of consensus modelsdeveloped for S. cerevisiae
| 模型版本 | 反应数 | 代谢物数 | 基因数 | 主要优化点 | 存在的问题 | 年份 | 参考文献 |
|---|---|---|---|---|---|---|---|
| Yeast1 | 1857 | 1168 | 832 | 第一代共识模型,统一代谢物注释 | 网络缺乏完整性、连通性,代谢反应覆盖率低 | 2008 | [ |
| Yeast4 | 2030 | 1481 | 924 | 增加脂质代谢反应,提高网络连通性 | 代谢反应覆盖率低 | 2010 | [ |
| Yeast5 | 2110 | 1655 | 918 | 增加鞘脂代谢反应,更新GPR关系 | 存在一定“阻塞反应”,引入假反应连通网络,不符合实际 | 2012 | [ |
| Yeast6 | 1888 | 1458 | 900 | 移除无明确功能或无参考来源的代谢物 | 模拟无氧生长存在问题,仍有一定“阻塞反应” | 2013 | [ |
| Yeast7 | 3493 | 2218 | 916 | 修正脂肪酸、甘油酯和甘油磷脂代谢,显著减少“阻塞反应”占比 | 基因数过少,无法充分整合多组学数据 | 2013 | [ |
| Yeast8 | 3949 | 2680 | 1133 | 对基因和反应进行大规模扩展,可引入蛋白质结构数据 | 反应质量和电荷不平衡,同工酶注释冗余,缺乏热力学约束 | 2019 | [ |
| Yeast9 | 4130 | 2805 | 1162 | 新增标准吉布斯自由能数据,提升反应准确性 | 无法精确表现细胞器代谢活动,多组学综合分析准确率较低 | 2024 | [ |
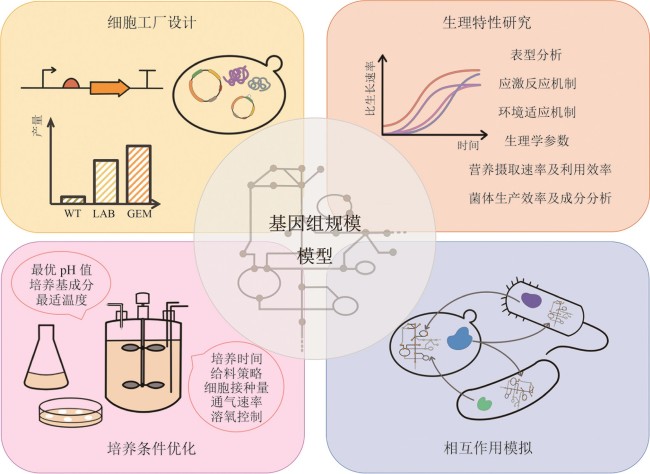
Fig. 2 Applications of the genome-scale models(The figure shows the applications of the genome-scale models in guiding cell factory design to enhance the yield of target products, assisting in the exploration of cellular physiological traits under different environments, optimizing cell culture conditions such as medium composition and temperature, and simulating metabolic exchanges and interactions within co-cultured microbial communities.)
| 1 | GOFFEAU A, BARRELL B G, BUSSEY H, et al. Life with 6000 genes[J]. Science, 1996, 274(5287): 546-567. |
| 2 | BERNARD A, ROSSIGNOL T, PARK Y K. Biotechnological approaches for producing natural pigments in yeasts[J]. Trends in Biotechnology, 2024, 42(12): 1644-1662. |
| 3 | LIAN J Z, MISHRA S, ZHAO H M. Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications[J]. Metabolic Engineering, 2018, 50: 85-108. |
| 4 | ZHANG Y W, YANG J J, QIAN F H, et al. Engineering a xylose fermenting yeast for lignocellulosic ethanol production[J]. Nature Chemical Biology, 2025, 21(3): 443-450. |
| 5 | IGUCHI H, YURIMOTO H, SAKAI Y. Interactions of methylotrophs with plants and other heterotrophic bacteria[J]. Microorganisms, 2015, 3(2): 137-151. |
| 6 | HENRIQUES D, MINEBOIS R, DOS SANTOS D, et al. A dynamic genome-scale model identifies metabolic pathways associated with cold tolerance in Saccharomyces kudriavzevii [J]. Microbiology Spectrum, 2023, 11(3): e03519-22. |
| 7 | HASSAN Y, CHEW S Y, THAN L T L. Candida glabrata: pathogenicity and resistance mechanisms for adaptation and survival[J]. Journal of Fungi, 2021, 7(8): 667. |
| 8 | BERNAUER L, RADKOHL A, LEHMAYER L G K, et al. Komagataella phaffii as emerging model organism in fundamental research[J]. Frontiers in Microbiology, 2021, 11: 607028. |
| 9 | CAVALLO E, CHARREAU H, CERRUTTI P, et al. Yarrowia lipolytica: a model yeast for citric acid production[J]. FEMS Yeast Research, 2017, 17(8): fox084. |
| 10 | MISHRA P, LEE N R, LAKSHMANAN M, et al. Genome-scale model-driven strain design for dicarboxylic acid production in Yarrowia lipolytica [J]. BMC Systems Biology, 2018, 12(S2): 12. |
| 11 | EDWARDS J S, PALSSON B O. Systems properties of the Haemophilus influenzae Rd metabolic genotype[J]. Journal of Biological Chemistry, 1999, 274(25): 17410-17416. |
| 12 | KANEHISA M, GOTO S. KEGG: kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Research, 2000, 28(1): 27-30. |
| 13 | KARP P D, RILEY M, SAIER M, et al. The EcoCyc and MetaCyc databases[J]. Nucleic Acids Research, 2000, 28(1): 56-59. |
| 14 | SCHAEFER C F, ANTHONY K, KRUPA S, et al. PID: the pathway interaction database[J]. Nucleic Acids Research, 2009, 37(Database issue): D674-D679. |
| 15 | HEIRENDT L, ARRECKX S, PFAU T, et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0[J]. Nature Protocols, 2019, 14(3): 639-702. |
| 16 | EBRAHIM A, LERMAN J A, PALSSON B O, et al. COBRApy: COnstraints-based reconstruction and analysis for python[J]. BMC Systems Biology, 2013, 7: 74. |
| 17 | WANG H, MARCIŠAUSKAS S, SÁNCHEZ B J, et al. RAVEN 2.0: a versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor [J]. PLoS Computational Biology, 2018, 14(10): e1006541. |
| 18 | BÜCHEL F, RODRIGUEZ N, SWAINSTON N, et al. Path2Models: large-scale generation of computational models from biochemical pathway maps[J]. BMC Systems Biology, 2013, 7: 116. |
| 19 | MACHADO D, ANDREJEV S, TRAMONTANO M, et al. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities[J]. Nucleic Acids Research, 2018, 46(15): 7542-7553. |
| 20 | HENRY C S, DEJONGH M, BEST A A, et al. High-throughput generation, optimization and analysis of genome-scale metabolic models[J]. Nature Biotechnology, 2010, 28(9): 977-982. |
| 21 | ARKIN A P, COTTINGHAM R W, HENRY C S, et al. KBase: the United States department of energy systems biology knowledgebase[J]. Nature Biotechnology, 2018, 36(7): 566-569. |
| 22 | KUEPFER L, SAUER U, BLANK L M. Metabolic functions of duplicate genes in Saccharomyces cerevisiae [J]. Genome Research, 2005, 15(10): 1421-1430. |
| 23 | MACKIE A, KESELER I M, NOLAN L, et al. Dead end metabolites: defining the known unknowns of the E. coli metabolic network[J]. PLoS One, 2013, 8(9): e75210. |
| 24 | ORTH J D, CONRAD T M, NA J, et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism: 2011[J]. Molecular Systems Biology, 2011, 7: 535. |
| 25 | KING Z A, LU J, DRÄGER A, et al. BiGG Models: a platform for integrating, standardizing and sharing genome-scale models[J]. Nucleic Acids Research, 2016, 44(D1): D515-D522. |
| 26 | CHELLIAH V, LAIBE C, NOVÈRE N L. BioModels database: a repository of mathematical models of biological processes[M/OL]//DUBITZKY W, WOLKENHAUER O, CHO K H, et al. Encyclopedia of systems biology. New York: Springer, 2013: 134-138. (2013-04-25)[2024-12-01]. . |
| 27 | ORTH J D, THIELE I, PALSSON B Ø. What is flux balance analysis?[J]. Nature Biotechnology, 2010, 28(3): 245-248. |
| 28 | MACGILLIVRAY M, KO A, GRUBER E, et al. Robust analysis of fluxes in genome-scale metabolic pathways[J]. Scientific Reports, 2017, 7(1): 268. |
| 29 | O'BRIEN E J, MONK J M, PALSSON B O. Using genome-scale models to predict biological capabilities[J]. Cell, 2015, 161(5): 971-987. |
| 30 | COVERT M W, XIAO N, CHEN T J, et al. Integrating metabolic, transcriptional regulatory and signal transduction models in Escherichia coli [J]. Bioinformatics, 2008, 24(18): 2044-2050. |
| 31 | SÁNCHEZ B J, ZHANG C, NILSSON A, et al. Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints[J]. Molecular Systems Biology, 2017, 13(8): 935. |
| 32 | THIELE I, JAMSHIDI N, FLEMING R M T, et al. Genome-scale reconstruction of Escherichia coli’s transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization[J]. PLoS Computational Biology, 2009, 5(3): e1000312. |
| 33 | PATIL K R, NIELSEN J. Uncovering transcriptional regulation of metabolism by using metabolic network topology[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(8): 2685-2689. |
| 34 | LU H Z, LI F R, SÁNCHEZ B J, et al. A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism[J]. Nature Communications, 2019, 10(1): 3586. |
| 35 | CHEN Y, GUSTAFSSON J, TAFUR RANGEL A, et al. Reconstruction, simulation and analysis of enzyme-constrained metabolic models using GECKO Toolbox 3.0[J]. Nature Protocols, 2024, 19(3): 629-667. |
| 36 | ADADI R, VOLKMER B, MILO R, et al. Prediction of microbial growth rate versus biomass yield by a metabolic network with kinetic parameters[J]. PLoS Computational Biology, 2012, 8(7): e1002575. |
| 37 | MAO Z T, ZHAO X, YANG X, et al. ECMpy, a simplified workflow for constructing enzymatic constrained metabolic network model[J]. Biomolecules, 2022, 12(1): 65. |
| 38 | YANG X, MAO Z T, ZHAO X, et al. Integrating thermodynamic and enzymatic constraints into genome-scale metabolic models[J]. Metabolic Engineering, 2021, 67: 133-144. |
| 39 | BI X Y, CHENG Y, XU X H, et al. etiBsu1209: a comprehensive multiscale metabolic model for Bacillus subtilis [J]. Biotechnology and Bioengineering, 2023, 120(6): 1623-1639. |
| 40 | LLOYD C J, EBRAHIM A, YANG L, et al. COBRAme: a computational framework for genome-scale models of metabolism and gene expression[J]. PLoS Computational Biology, 2018, 14(7): e1006302. |
| 41 | BULOVIĆ A, FISCHER S, DINH M, et al. Automated generation of bacterial resource allocation models[J]. Metabolic Engineering, 2019, 55: 12-22. |
| 42 | OFTADEH O, SALVY P, MASID M, et al. A genome-scale metabolic model of Saccharomyces cerevisiae that integrates expression constraints and reaction thermodynamics[J]. Nature Communications, 2021, 12(1): 4790. |
| 43 | JESKE L, PLACZEK S, SCHOMBURG I, et al. BRENDA in 2019: a European ELIXIR core data resource[J]. Nucleic Acids Research, 2019, 47(D1): D542-D549. |
| 44 | LI F R, YUAN L, LU H Z, et al. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction[J]. Nature Catalysis, 2022, 5(8): 662-672. |
| 45 | O'BRIEN E J, LERMAN J A, CHANG R L, et al. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction[J]. Molecular Systems Biology, 2013, 9: 693. |
| 46 | FANG X, LLOYD C J, PALSSON B O. Reconstructing organisms in silico: genome-scale models and their emerging applications[J]. Nature Reviews Microbiology, 2020, 18(12): 731-743. |
| 47 | MAN O, PILPEL Y. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species[J]. Nature Genetics, 2007, 39(3): 415-421. |
| 48 | DUARTE N C, BECKER S A, JAMSHIDI N, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(6): 1777-1782. |
| 49 | VANDERWAEREN L, DOK R, VOORDECKERS K, et al. Saccharomyces cerevisiae as a model system for eukaryotic cell biology, from cell cycle control to DNA damage response[J]. International Journal of Molecular Sciences, 2022, 23(19): 11665. |
| 50 | FÖRSTER J, FAMILI I, FU P, et al. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network[J]. Genome Research, 2003, 13(2): 244-253. |
| 51 | DUARTE N C, HERRGÅRD M J, PALSSON B Ø. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model[J]. Genome Research, 2004, 14(7): 1298-1309. |
| 52 | NOOKAEW I, JEWETT M C, MEECHAI A, et al. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism[J]. BMC Systems Biology, 2008, 2: 71. |
| 53 | HERRGÅRD M J, SWAINSTON N, DOBSON P, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology[J]. Nature Biotechnology, 2008, 26(10): 1155-1160. |
| 54 | ZOMORRODI A R, MARANAS C D. Improving the iMM904 S. cerevisiae metabolic model using essentiality and synthetic lethality data[J]. BMC Systems Biology, 2010, 4: 178. |
| 55 | HEAVNER B D, SMALLBONE K, BARKER B, et al. Yeast5-an expanded reconstruction of the Saccharomyces cerevisiae metabolic network[J]. BMC Systems Biology, 2012, 6: 55. |
| 56 | ÖSTERLUND T, NOOKAEW I, BORDEL S, et al. Mapping condition-dependent regulation of metabolism in yeast through genome-scale modeling[J]. BMC Systems Biology, 2013, 7: 36. |
| 57 | ZHANG C Y, SÁNCHEZ B J, LI F R, et al. Yeast9: a consensus genome-scale metabolic model for S. cerevisiae curated by the community[J]. Molecular Systems Biology, 2024, 20(10): 1134-1150. |
| 58 | DOBSON P D, SMALLBONE K, JAMESON D, et al. Further developments towards a genome-scale metabolic model of yeast[J]. BMC Systems Biology, 2010, 4: 145. |
| 59 | HEAVNER B D, SMALLBONE K, PRICE N D, et al. Version 6 of the consensus yeast metabolic network refines biochemical coverage and improves model performance[J]. Database, 2013, 2013: bat059. |
| 60 | AUNG H W, HENRY S A, WALKER L P. Revising the representation of fatty acid, glycerolipid, and glycerophospholipid metabolism in the consensus model of yeast metabolism[J]. Industrial Biotechnology, 2013, 9(4): 215-228. |
| 61 | HAN S Y, WU K, WANG Y H, et al. Auxotrophy-based curation improves the consensus genome-scale metabolic model of yeast[J]. Synthetic and Systems Biotechnology, 2024, 9(4): 861-870. |
| 62 | MO M L, PALSSON B Ø, HERRGÅRD M J. Connecting extracellular metabolomic measurements to intracellular flux states in yeast[J]. BMC Systems Biology, 2009, 3: 37. |
| 63 | TSOUKA S, HATZIMANIKATIS V. redLips: a comprehensive mechanistic model of the lipid metabolic network of yeast[J]. FEMS Yeast Research, 2020, 20(2): foaa006. |
| 64 | SCOTT W T JR, SMID E J, NOTEBAART R A, et al. Curation and analysis of a Saccharomyces cerevisiae genome-scale metabolic model for predicting production of sensory impact molecules under enological conditions[J]. Processes, 2020, 8(9): 1195. |
| 65 | CHOWDHURY R, CHOWDHURY A, MARANAS C D. Using gene essentiality and synthetic lethality information to correct yeast and CHO cell genome-scale models[J]. Metabolites, 2015, 5(4): 536-570. |
| 66 | ATA Ö, ERGÜN B G, FICKERS P, et al. What makes Komagataella phaffii non-conventional?[J]. FEMS Yeast Research, 2021, 21(8): foab059. |
| 67 | SOHN S B, GRAF A B, KIM T Y, et al. Genome-scale metabolic model of methylotrophic yeast Pichia pastoris and its use for in silico analysis of heterologous protein production[J]. Biotechnology Journal, 2010, 5(7): 705-715. |
| 68 | CHUNG B K, SELVARASU S, CAMATTARI A, et al. Genome-scale metabolic reconstruction and in silico analysis of methylotrophic yeast Pichia pastoris for strain improvement[J]. Microbial Cell Factories, 2010, 9: 50. |
| 69 | CASPETA L, SHOAIE S, AGREN R, et al. Genome-scale metabolic reconstructions of Pichia stipitis and Pichia pastoris and in silico evaluation of their potentials[J]. BMC Systems Biology, 2012, 6: 24. |
| 70 | TOMÀS-GAMISANS M, FERRER P, ALBIOL J. Integration and validation of the genome-scale metabolic models of Pichia pastoris: a comprehensive update of protein glycosylation pathways, lipid and energy metabolism[J]. PLoS One, 2016, 11(1): e0148031. |
| 71 | TOMÀS-GAMISANS M, FERRER P, ALBIOL J. Fine-tuning the P. pastoris iMT1026 genome-scale metabolic model for improved prediction of growth on methanol or glycerol as sole carbon sources[J]. Microbial Biotechnology, 2018, 11(1): 224-237. |
| 72 | LOIRA N, DULERMO T, NICAUD J M, et al. A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica [J]. BMC Systems Biology, 2012, 6: 35. |
| 73 | KERKHOVEN E J, POMRANING K R, BAKER S E, et al. Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica [J]. NPJ Systems Biology and Applications, 2016, 2: 16005. |
| 74 | GUO Y F, SU L Q, LIU Q, et al. Dissecting carbon metabolism of Yarrowia lipolytica type strain W29 using genome-scale metabolic modelling[J]. Computational and Structural Biotechnology Journal, 2022, 20: 2503-2511. |
| 75 | ZHU Z W, ZHANG S F, LIU H W, et al. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides [J]. Nature Communications, 2012, 3: 1112. |
| 76 | TIUKOVA I A, PRIGENT S, NIELSEN J, et al. Genome-scale model of Rhodotorula toruloides metabolism[J]. Biotechnology and Bioengineering, 2019, 116(12): 3396-3408. |
| 77 | KIM J H, CORADETTI S T, KIM Y M, et al. Multi-omics driven metabolic network reconstruction and analysis of lignocellulosic carbon utilization in Rhodosporidium toruloides [J]. Frontiers in Bioengineering and Biotechnology, 2021, 8: 612832. |
| 78 | LIEBAL U W, FABRY B A, RAVIKRISHNAN A, et al. Genome-scale model reconstruction of the methylotrophic yeast Ogataea polymorpha [J]. BMC Biotechnology, 2021, 21(1): 23. |
| 79 | MISHRA P, PARK G Y, LAKSHMANAN M, et al. Genome-scale metabolic modeling and in silico analysis of lipid accumulating yeast Candida tropicalis for dicarboxylic acid production[J]. Biotechnology and Bioengineering, 2016, 113(9): 1993-2004. |
| 80 | XU N, LIU L M, ZOU W, et al. Reconstruction and analysis of the genome-scale metabolic network of Candida glabrata [J]. Molecular BioSystems, 2013, 9(2): 205-216. |
| 81 | VYAS A, FREITAS A V, RALSTON Z A, et al. Fission yeast Schizosaccharomyces pombe: a unicellular "micromammal" model organism[J]. Current Protocols, 2021, 1(6): e151. |
| 82 | SOHN S B, KIM T Y, LEE J H, et al. Genome-scale metabolic model of the fission yeast Schizosaccharomyces pombe and the reconciliation of in silico/in vivo mutant growth[J]. BMC Systems Biology, 2012, 6: 49. |
| 83 | GRIGAITIS P, GRUNDEL D A J, VAN PELT-KLEINJAN E, et al. A computational toolbox to investigate the metabolic potential and resource allocation in fission yeast[J]. mSystems, 2022, 7(4): e0042322. |
| 84 | SPOHNER S C, SCHAUM V, QUITMANN H, et al. Kluyveromyces lactis: an emerging tool in biotechnology[J]. Journal of Biotechnology, 2016, 222: 104-116. |
| 85 | DIAS O, PEREIRA R, GOMBERT A K, et al. iOD907, the first genome-scale metabolic model for the milk yeast Kluyveromyces lactis [J]. Biotechnology Journal, 2014, 9(6): 776-790. |
| 86 | MARCIŠAUSKAS S, JI B Y, NIELSEN J. Reconstruction and analysis of a Kluyveromyces marxianus genome-scale metabolic model[J]. BMC Bioinformatics, 2019, 20(1): 551. |
| 87 | PAPINI M, NOOKAEW I, UHLÉN M, et al. Scheffersomyces stipitis: a comparative systems biology study with the Crabtree positive yeast Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2012, 11: 136. |
| 88 | BALAGURUNATHAN B, JONNALAGADDA S, TAN L, et al. Reconstruction and analysis of a genome-scale metabolic model for Scheffersomyces stipitis [J]. Microbial Cell Factories, 2012, 11: 27. |
| 89 | HILLIARD M, DAMIANI A, HE Q P, et al. Elucidating redox balance shift in Scheffersomyces stipitis’ fermentative metabolism using a modified genome-scale metabolic model[J]. Microbial Cell Factories, 2018, 17(1): 140. |
| 90 | LI G, HU Y T, ZRIMEC J, et al. Bayesian genome scale modelling identifies thermal determinants of yeast metabolism[J]. Nature Communications, 2021, 12(1): 190. |
| 91 | CHEN Y, LI F R, MAO J W, et al. Yeast optimizes metal utilization based on metabolic network and enzyme kinetics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(12): e2020154118. |
| 92 | ELSEMMAN I E, RODRIGUEZ PRADO A, GRIGAITIS P, et al. Whole-cell modeling in yeast predicts compartment-specific proteome constraints that drive metabolic strategies[J]. Nature Communications, 2022, 13(1): 801. |
| 93 | LI F R, CHEN Y, QI Q, et al. Improving recombinant protein production by yeast through genome-scale modeling using proteome constraints[J]. Nature Communications, 2022, 13(1): 2969. |
| 94 | DOMENZAIN I, SÁNCHEZ B, ANTON M, et al. Reconstruction of a catalogue of genome-scale metabolic models with enzymatic constraints using GECKO 2.0[J]. Nature Communications, 2022, 13(1): 3766. |
| 95 | REĶĒNA A, PINHEIRO M J, BONTURI N, et al. Genome-scale metabolic modeling reveals metabolic trade-offs associated with lipid production in Rhodotorula toruloides [J]. PLoS Computational Biology, 2023, 19(4): e1011009. |
| 96 | BATTJES J, GRIGAITIS P, HOVING M, et al. Mitochondrial efficiency determines Crabtree effect across yeasts[EB/OL]. bioRxiv, 2024: 2024.11.01.621473. (2024-11-03)[2024-11-15]. . |
| 97 | GRIGAITIS P, VAN DEN BOGAARD S L, TEUSINK B. Elevated energy costs of biomass production in mitochondrial respiration-deficient Saccharomyces cerevisia [J]. FEMS Yeast Research, 2023, 23: foad008. |
| 98 | CHOI B, TAFUR RANGEL A, KERKHOVEN E J, et al. Engineering of Saccharomyces cerevisiae for enhanced metabolic robustness and L-lactic acid production from lignocellulosic biomass[J]. Metabolic Engineering, 2024, 84: 23-33. |
| 99 | QIN N, LI L Y, WAN X Z, et al. Increased CO2 fixation enables high carbon-yield production of 3-hydroxypropionic acid in yeast[J]. Nature Communications, 2024, 15(1): 1591. |
| 100 | ISHCHUK O P, DOMENZAIN I, SÁNCHEZ B J, et al. Genome-scale modeling drives 70-fold improvement of intracellular heme production in Saccharomyces cerevisiae [J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(30): e2108245119. |
| 101 | BERNAT-CAMPS N, EBNER K, SCHUSTERBAUER V, et al. Enabling growth-decoupled Komagataella phaffii recombinant protein production based on the methanol-free PDH promoter[J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1130583. |
| 102 | WEFELMEIER K, SCHMITZ S, KÖSTERS B J, et al. Methanol bioconversion into C3, C4, and C5 platform chemicals by the yeast Ogataea polymorpha [J]. Microbial Cell Factories, 2024, 23(1): 8. |
| 103 | VEIGA MOREIRA J DA, JOLICOEUR M, SCHWARTZ L, et al. Fine-tuning mitochondrial activity in Yarrowia lipolytica for citrate overproduction[J]. Scientific Reports, 2021, 11(1): 878. |
| 104 | SCOTT W T JR, SMID E J, BLOCK D E, et al. Metabolic flux sampling predicts strain-dependent differences related to aroma production among commercial wine yeasts[J]. Microbial Cell Factories, 2021, 20(1): 204. |
| 105 | MESQUITA T J B, SARGO C R, FUZER NETO J R, et al. Metabolic fluxes-oriented control of bioreactors: a novel approach to tune micro-aeration and substrate feeding in fermentations[J]. Microbial Cell Factories, 2019, 18(1): 150. |
| 106 | BOOJARI M A, GHALEDARI F R, MOTAMEDIAN E, et al. Developing a metabolic model-based fed-batch feeding strategy for Pichia pastoris fermentation through fine-tuning of the methanol utilization pathway[J]. Microbial Biotechnology, 2023, 16(6): 1344-1359. |
| 107 | TAMIRES MOREIRA MELO N, PONTES G C, PROCÓPIO D P, et al. Evaluation of product distribution in chemostat and batch fermentation in lactic acid-producing Komagataella phaffii strains utilizing glycerol as substrate[J]. Microorganisms, 2020, 8(5): 781. |
| 108 | FERNÁNDEZ-NIÑO M, RODRÍGUEZ-CUBILLOS M J, HERRERA-ROCHA F, et al. Dissecting industrial fermentations of fine flavour cocoa through metagenomic analysis[J]. Scientific Reports, 2021, 11(1): 8638. |
| 109 | PONOMAROVA O, GABRIELLI N, SÉVIN D C, et al. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow[J]. Cell Systems, 2017, 5(4): 345-357.e6. |
| 110 | ÖSTERLUND T, NOOKAEW I, NIELSEN J. Fifteen years of large scale metabolic modeling of yeast: developments and impacts[J]. Biotechnology Advances, 2012, 30(5): 979-988. |
| 111 | GU C D, KIM G B, KIM W J, et al. Current status and applications of genome-scale metabolic models[J]. Genome Biology, 2019, 20(1): 121. |
| 112 | DOMENZAIN I, LI F R, KERKHOVEN E J, et al. Evaluating accessibility, usability and interoperability of genome-scale metabolic models for diverse yeasts species[J]. FEMS Yeast Research, 2021, 21(1): foab002. |
| 113 | MAHADEVAN R, LOVLEY D R. The degree of redundancy in metabolic genes is linked to mode of metabolism[J]. Biophysical Journal, 2008, 94(4): 1216-1220. |
| 114 | DEUTSCHER D, MEILIJSON I, KUPIEC M, et al. Multiple knockout analysis of genetic robustness in the yeast metabolic network[J]. Nature Genetics, 2006, 38(9): 993-998. |
| 115 | CHEN L F, VITKUP D. Predicting genes for orphan metabolic activities using phylogenetic profiles[J]. Genome Biology, 2006, 7(2): R17. |
| 116 | RAZAGHI-MOGHADAM Z, BABADI F S, NIKOLOSKI Z. Harnessing the optimization of enzyme catalytic rates in engineering of metabolic phenotypes[J]. PLoS Computational Biology, 2024, 20(11): e1012576. |
| 117 | SÁNCHEZ B J, LI F R, KERKHOVEN E J, et al. SLIMEr: probing flexibility of lipid metabolism in yeast with an improved constraint-based modeling framework[J]. BMC Systems Biology, 2019, 13(1): 4. |
| 118 | DE MOURA FERREIRA M A, WENDERING P, AREND M, et al. Accurate prediction of in vivo protein abundances by coupling constraint-based modelling and machine learning[J]. Metabolic Engineering, 2023, 80: 184-192. |
| 119 | BURGARD A P, NIKOLAEV E V, SCHILLING C H, et al. Flux coupling analysis of genome-scale metabolic network reconstructions[J]. Genome Research, 2004, 14(2): 301-312. |
| 120 | SMALLBONE K, SIMEONIDIS E, SWAINSTON N, et al. Towards a genome-scale kinetic model of cellular metabolism[J]. BMC Systems Biology, 2010, 4: 6. |
| 121 | MOHAMMADIPEYHANI H, HAFNER J, SVESHNIKOVA A, et al. Expanding biochemical knowledge and illuminating metabolic dark matter with ATLASx[J]. Nature Communications, 2022, 13(1): 1560. |
| 122 | NIELSEN J, KEASLING J D. Engineering cellular metabolism[J]. Cell, 2016, 164(6): 1185-1197. |
| 123 | CROWE S A, LIU Y Z, ZHAO X X, et al. Advances in engineering nucleotide sugar metabolism for natural product glycosylation in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2024, 13(6): 1589-1599. |
| 124 | LERMA-ORTIZ C, JEFFRYES J G, COOPER A J L, et al. ‘Nothing of chemistry disappears in biology’: the top 30 damage-prone endogenous metabolites[J]. Biochemical Society Transactions, 2016, 44(3): 961-971. |
| 125 | WU K, LIU H, SUN M, et al. Yeast-MetaTwin for systematically exploring yeast metabolism through retrobiosynthesis and deep learning[EB/OL]. bioRxiv, 2024: 2024.09.02.610684. (2024-09-02)[2024-11-01]. . |
| 126 | NIELSEN J C, NIELSEN J. Development of fungal cell factories for the production of secondary metabolites: Linking genomics and metabolism[J]. Synthetic and Systems Biotechnology, 2017, 2(1): 5-12. |
| 127 | LU H Z, KERKHOVEN E J, NIELSEN J. Multiscale models quantifying yeast physiology: towards a whole-cell model[J]. Trends in Biotechnology, 2022, 40(3): 291-305. |
| 128 | SEN P, OREŠIČ M. Integrating omics data in genome-scale metabolic modeling: a methodological perspective for precision medicine[J]. Metabolites, 2023, 13(7): 855. |
| 129 | GUO J, IBANEZ-LOPEZ A S, GAO H Y, et al. Automated chemical reaction extraction from scientific literature[J]. Journal of Chemical Information and Modeling, 2022, 62(9): 2035-2045. |
| 130 | BESSELL B, LOECKER J, ZHAO Z Y, et al. COMO: a pipeline for multi-omics data integration in metabolic modeling and drug discovery[J]. Briefings in Bioinformatics, 2023, 24(6): bbad387. |
| 131 | THIELE I, PALSSON B Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction[J]. Nature Protocols, 2010, 5(1): 93-121. |
| [1] | ZHANG Yiqing, LIU Gaowen. Exploration of gene functions and library construction for engineering strains from a synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(3): 685-700. |
| [2] | GAO Qi, XIAO Wenhai. Advances in the biosynthesis of monoterpenes by yeast [J]. Synthetic Biology Journal, 2025, 6(2): 357-372. |
| [3] | SHENG Zhouhuang, CHEN Zhixian, ZHANG Yan. Research progress of yeast mannoproteins [J]. Synthetic Biology Journal, 2025, 6(2): 408-421. |
| [4] | XIAO Sen, HU Litao, SHI Zhicheng, WANG Fayin, YU Siting, DU Guocheng, CHEN Jian, KANG Zhen. Research advances in biosynthesis of hyaluronic acid with controlled molecular weights [J]. Synthetic Biology Journal, 2025, 6(2): 445-460. |
| [5] | WANG Qian, GUO Shiting, XIN Bo, ZHONG Cheng, WANG Yu. Advances in biosynthesis of L-arginine using engineered microorganisms [J]. Synthetic Biology Journal, 2025, 6(2): 290-305. |
| [6] | ZUO Yimeng, ZHANG Jiaojiao, LIAN Jiazhang. Enabling technology for the biosynthesis of cosmetic raw materials with Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2025, 6(2): 233-253. |
| [7] | HUANG Shuhan, MA He, LUO Yunzi. Research progress in the biosynthesis of salidroside [J]. Synthetic Biology Journal, 2025, 6(2): 391-407. |
| [8] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [11] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [12] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [13] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||