Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (5): 1211-1226.DOI: 10.12211/2096-8280.2024-024
• Invited Review • Previous Articles
Reprogramming microbial chassis for low-cost bioprodcution of tailor-made polyhydroxyalkanoates
- 1.School of Life Sciences,Center for Synthetic and Systems Biology (CSSB),Department of Chemical Engineering,Tsinghua University,Beijing 100084,China
2.Key Laboratory of Biomedical Information Engineering of Ministry of Education,School of Life Science and Technology,Xi'an Jiaotong University,Xi'an 710049,Shaanxi,China
-
Received:2024-03-19Revised:2024-06-08Online:2024-11-20Published:2024-10-31 -
Contact:CHEN Guo-Qiang
重编程微生物底盘用于PHA材料的定制化低成本生物合成
- 1.清华大学生命科学学院,化学工程系,合成与系统生物学中心,北京 100084
2.西安交通大学生命科学与技术学院,生物医学信息工程教育部重点实验室,陕西 西安 710049
-
通讯作者:陈国强 -
作者简介:陈国强 (1963—),男,教授,博士生导师。研究方向为生物合成PHA材料及其下一代工业生物技术。 E-mail:chengq@mail.tsinghua.edu.cn -
基金资助:国家自然科学基金(32130001);国家重点研发计划“合成生物学”重点专项(2018YFA0900200)
CLC Number:
Cite this article
CHEN Guo-Qiang, TAN Dan. Reprogramming microbial chassis for low-cost bioprodcution of tailor-made polyhydroxyalkanoates[J]. Synthetic Biology Journal, 2024, 5(5): 1211-1226.
陈国强, 谭丹. 重编程微生物底盘用于PHA材料的定制化低成本生物合成[J]. 合成生物学, 2024, 5(5): 1211-1226.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-024
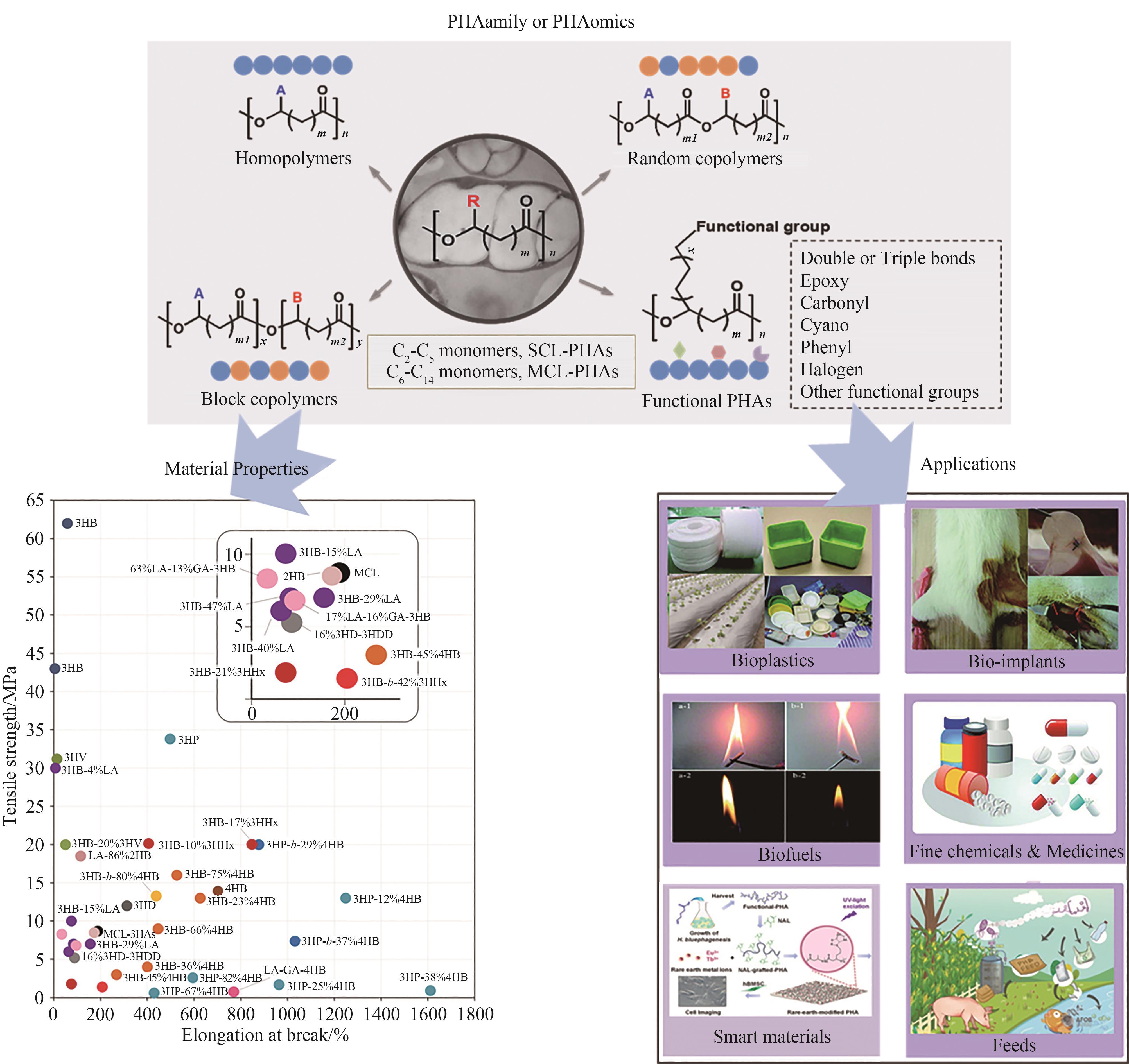
聚合物类型 Polymer type | 熔融温度(Tm)/℃ Melting temperature/℃ | 玻璃化转变温度(Tg)/℃ Glass transition temperature/℃ | 拉伸强度/MPa Tensile strength/MPa | 断裂伸长率/% Elongation at break/% |
|---|---|---|---|---|
| PHB | 178 | 4 | 43 | 5 |
| P(3HB-20% 3HV) | 145 | -1 | 20 | 50 |
| P(3HB-17% 3HHx) | 120 | -2 | 20 | 850 |
| P(4HB) | 58 | -48 | 104 | 1000 |
| P(3HB-45% 4HB) | 162 | -16 | 3 | 268 |
| P(3HP) | 78.1 | -17.9 | 33.8 | 497.6 |
| P(7% 3HHx-3HO) | 61 | -37.8 | 7.4 | 346.3 |
| P(10% 3HHx-86% 3HO-4% 3HD) | 61 | -35 | 10 | 300 |
| PP | 186 | -10 | 38 | 400 |
| PET | 262 | — | 56 | 8300 |
| HDPE | 135 | — | 29 | — |
Table 1 Comparison of material properties between typical PHAs and traditional plastics [12]
聚合物类型 Polymer type | 熔融温度(Tm)/℃ Melting temperature/℃ | 玻璃化转变温度(Tg)/℃ Glass transition temperature/℃ | 拉伸强度/MPa Tensile strength/MPa | 断裂伸长率/% Elongation at break/% |
|---|---|---|---|---|
| PHB | 178 | 4 | 43 | 5 |
| P(3HB-20% 3HV) | 145 | -1 | 20 | 50 |
| P(3HB-17% 3HHx) | 120 | -2 | 20 | 850 |
| P(4HB) | 58 | -48 | 104 | 1000 |
| P(3HB-45% 4HB) | 162 | -16 | 3 | 268 |
| P(3HP) | 78.1 | -17.9 | 33.8 | 497.6 |
| P(7% 3HHx-3HO) | 61 | -37.8 | 7.4 | 346.3 |
| P(10% 3HHx-86% 3HO-4% 3HD) | 61 | -35 | 10 | 300 |
| PP | 186 | -10 | 38 | 400 |
| PET | 262 | — | 56 | 8300 |
| HDPE | 135 | — | 29 | — |
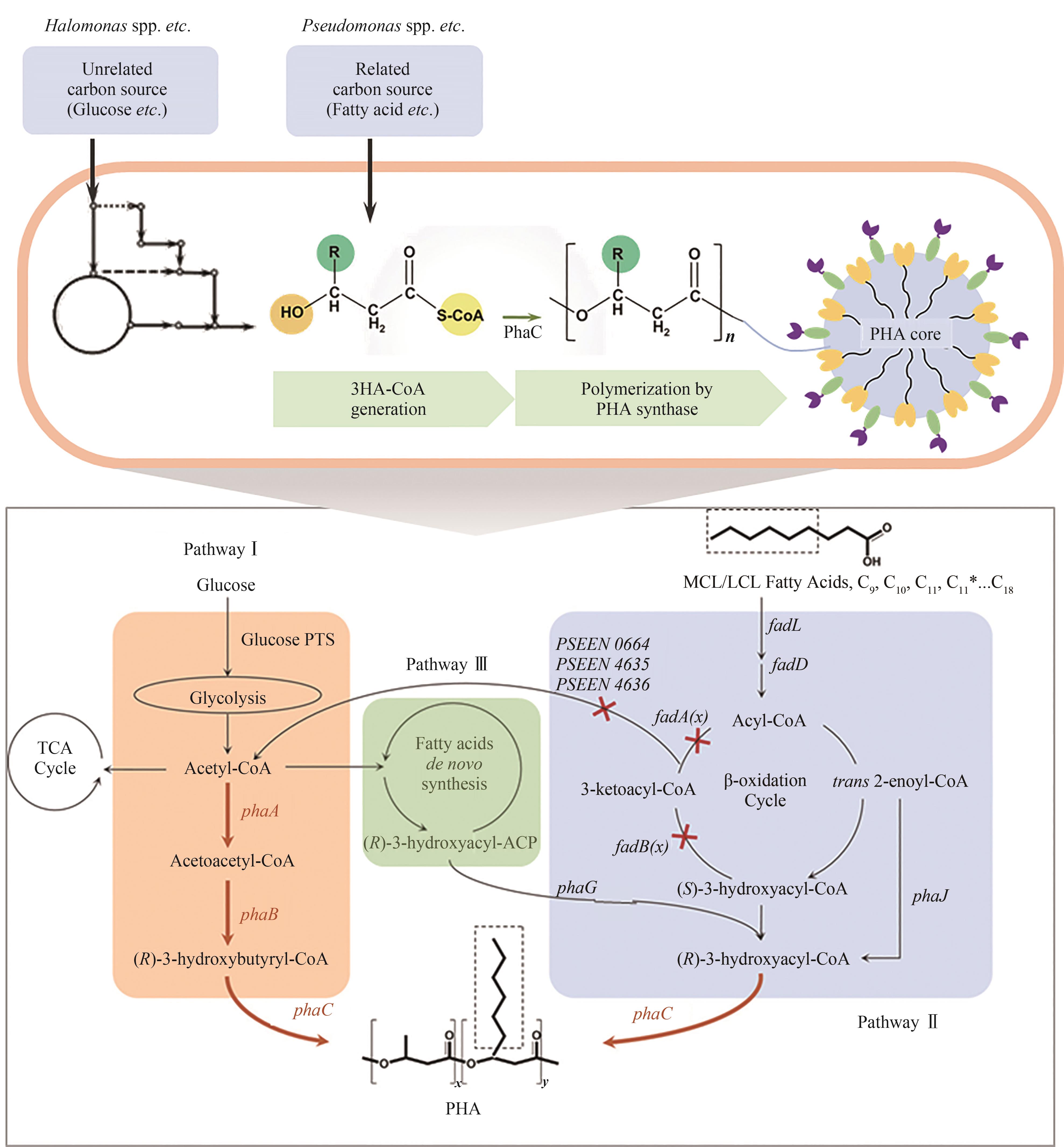
Fig. 2 The major PHA biosynthetic pathways(PHA biosynthetic pathways fed with related and unrelated carbon sources in the three PHA classic pathways: glucose glycolysis pathway, β-oxidation pathway, and de novo synthesis pathway offatty acids)
生产菌 Producers | PHA类型 PHAs | 底物 Substrates | 细胞干重/(g/L) Cell dry weight/(g/L) | PHA质量 分数/% PHA content/% | 最高体积产率/[g/(L·h)] Highest volumetric productivity/[g/(L·h)] | 参考文献 References |
|---|---|---|---|---|---|---|
| Escherichia coli | Various PHAs | Glucose | 141.6 | 73 | 4.63 | [ |
| Ralstonia eutropha | SCL-PHAs, MCL-PHAs, PHBHHx | Glucose Fatty acid | 232 | 80 | 3.14 | [ |
| Aeromonas hydrophila | PHBHHx | Fatty acid | 43.3 | 45.2 | 1.01 | [ |
| Pseudomonas spp. | MCL-PHAs | Fatty acid | 72.6 | 51.4 | 1.91 | [ |
| Halomonas spp. | SCL-PHAs | Glucose | 100 | 60~92 | 1.67~3.2 | [ |
Table 2 Main PHA producers and their maximal PHA yields
生产菌 Producers | PHA类型 PHAs | 底物 Substrates | 细胞干重/(g/L) Cell dry weight/(g/L) | PHA质量 分数/% PHA content/% | 最高体积产率/[g/(L·h)] Highest volumetric productivity/[g/(L·h)] | 参考文献 References |
|---|---|---|---|---|---|---|
| Escherichia coli | Various PHAs | Glucose | 141.6 | 73 | 4.63 | [ |
| Ralstonia eutropha | SCL-PHAs, MCL-PHAs, PHBHHx | Glucose Fatty acid | 232 | 80 | 3.14 | [ |
| Aeromonas hydrophila | PHBHHx | Fatty acid | 43.3 | 45.2 | 1.01 | [ |
| Pseudomonas spp. | MCL-PHAs | Fatty acid | 72.6 | 51.4 | 1.91 | [ |
| Halomonas spp. | SCL-PHAs | Glucose | 100 | 60~92 | 1.67~3.2 | [ |
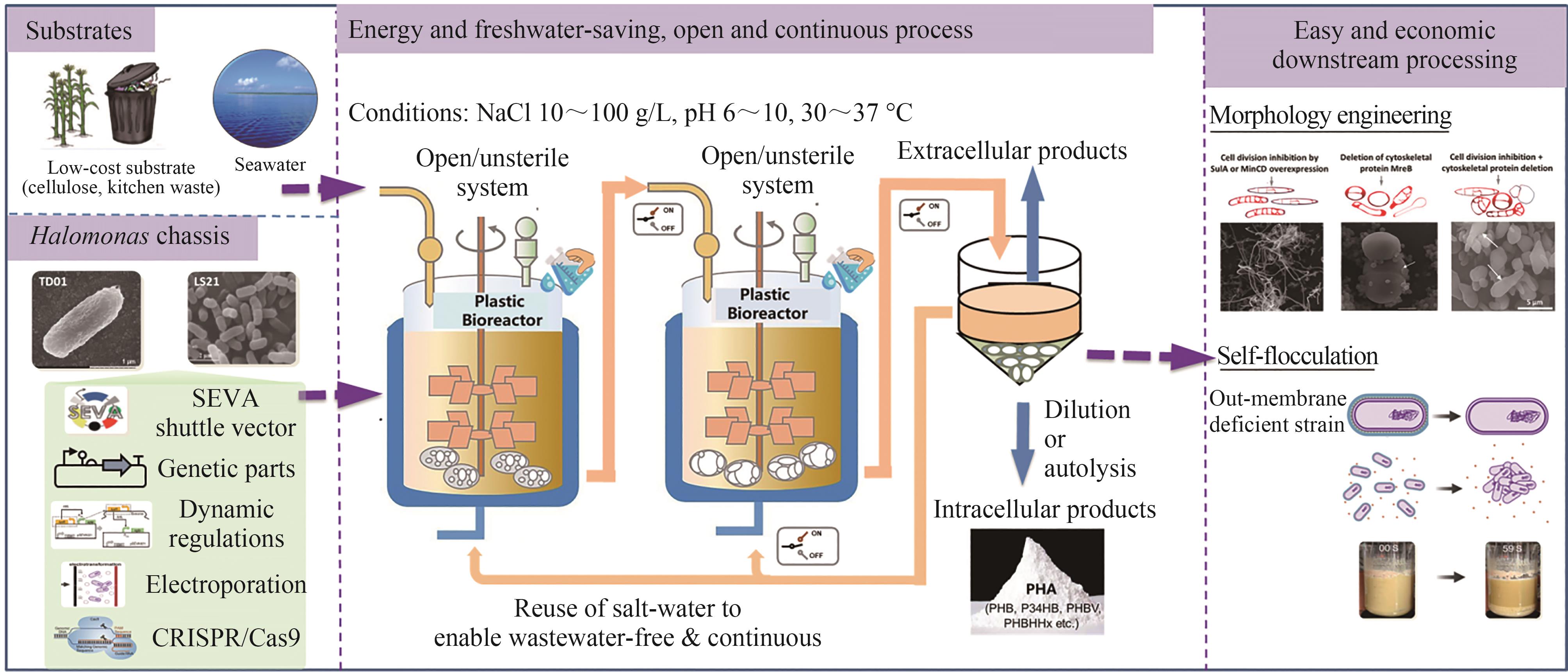
Fig. 3 Next Generation Industrial Biotechnology (NGIB) based on Halomonas spp.[80](An open/unsterile and continuous, energy and freshwater-saving bio-process for the production of various intracellular and extracellular products, which allows morphology engineering for the easy and economic downstream processing.)
公司 Companies | PHA类型 PHAs Types | 技术 Technology | 规模/(吨/年) Scale/(t/a) | 官方网站 Websites |
|---|---|---|---|---|
| Go!PHA, The Netherlands | All Types | PHA Global Promotion | Unknown | gopha.org |
| PhaBuilder, China | All Types | Halomonas spp. (NGIB①) | 1000~10 000 | www.phabuilder.com |
| Medpha, China | P3HB4HB | Halomonas spp. (NGIB①) | 100 | www.medpha.com.cn |
| COFCO, China | PHB | Halomonas spp. (NGIB①) | 1000 | www.cofco.com |
| Bluepha, China | PHBHHx | Ralstonia eutropha and NGIB | 1000 | www.bluepha.com |
| TianAn Biopolymer, China | PHBV | Ralstonia eutropha | 2000 | www.tianan-enmat.com |
| GreenBio, Tianjin, China | P3HB4HB | Escherichia coli | 10 000 | www.tjgreenbio.com |
| Ecomann, Shenzhen, China | P3HB4HB | Escherichia coli | 10 000 | ecomannbruce.plasway.com |
| RWDC, Singapore and USA | PHBHHx | Ralstonia eutropha | Unknown | www.rwdc-industries.com |
| Danimer Scientific, USA | PHBHHx | Ralstonia eutropha | 10 000 | danimerscientific.com |
| Full Cycle, USA | PHA② | non-GMO bacteria | Unknown | fullcyclebioplastics.com |
| Newlight, USA | PHB | Ocean microbes grown on greenhouse gas | Unknown | www.newlight.com |
| Metabolix, USA | P3HB4HB | Escherichia coli | 5000 | IP sold to CJ, Korea |
| BOSK Bioproducts, Canada | PHA② | Forest wastes for PHA production | Unknown | www.bosk-bioproducts.com |
| Genecis, Canada | PHBV | Unknown | Unknown | genecis.co |
| TerraVerdae Bioworks,Canada | PHA② | Unknown | Unknown | terraverdae.com |
| Kaneka, Japan | PHBHHx | Ralstonia eutropha | 5000 | www.kaneka.be |
| Nafigate, France | PHB | Toxic waste as substrates | Unknown | www.nafigate.com |
| CJ, Korea | P3HB4HB | Escherichia coli | Unknown | www.cj.co.kr |
| Helian Polymers, The Netherlands | PHB/PHBV | non-GMO bacteria | Unknown | helianpolymers.com |
| Biocycle, Brazil | PHB | Bacillus spp. | 100 | fapesp.br |
| Biomer, Germany | PHB | Alcaligenes latus | Unknown | biomer.de |
| Bioextrax, Sweden | PHA② | Bioextrax DSP method | Unknown | bioextrax.com |
| SABIO srl, Italy | PHA② | Organic wastes for PHA production | Unknown | www.bio-on.it |
Table 3 PHAs commercialization companies[6]
公司 Companies | PHA类型 PHAs Types | 技术 Technology | 规模/(吨/年) Scale/(t/a) | 官方网站 Websites |
|---|---|---|---|---|
| Go!PHA, The Netherlands | All Types | PHA Global Promotion | Unknown | gopha.org |
| PhaBuilder, China | All Types | Halomonas spp. (NGIB①) | 1000~10 000 | www.phabuilder.com |
| Medpha, China | P3HB4HB | Halomonas spp. (NGIB①) | 100 | www.medpha.com.cn |
| COFCO, China | PHB | Halomonas spp. (NGIB①) | 1000 | www.cofco.com |
| Bluepha, China | PHBHHx | Ralstonia eutropha and NGIB | 1000 | www.bluepha.com |
| TianAn Biopolymer, China | PHBV | Ralstonia eutropha | 2000 | www.tianan-enmat.com |
| GreenBio, Tianjin, China | P3HB4HB | Escherichia coli | 10 000 | www.tjgreenbio.com |
| Ecomann, Shenzhen, China | P3HB4HB | Escherichia coli | 10 000 | ecomannbruce.plasway.com |
| RWDC, Singapore and USA | PHBHHx | Ralstonia eutropha | Unknown | www.rwdc-industries.com |
| Danimer Scientific, USA | PHBHHx | Ralstonia eutropha | 10 000 | danimerscientific.com |
| Full Cycle, USA | PHA② | non-GMO bacteria | Unknown | fullcyclebioplastics.com |
| Newlight, USA | PHB | Ocean microbes grown on greenhouse gas | Unknown | www.newlight.com |
| Metabolix, USA | P3HB4HB | Escherichia coli | 5000 | IP sold to CJ, Korea |
| BOSK Bioproducts, Canada | PHA② | Forest wastes for PHA production | Unknown | www.bosk-bioproducts.com |
| Genecis, Canada | PHBV | Unknown | Unknown | genecis.co |
| TerraVerdae Bioworks,Canada | PHA② | Unknown | Unknown | terraverdae.com |
| Kaneka, Japan | PHBHHx | Ralstonia eutropha | 5000 | www.kaneka.be |
| Nafigate, France | PHB | Toxic waste as substrates | Unknown | www.nafigate.com |
| CJ, Korea | P3HB4HB | Escherichia coli | Unknown | www.cj.co.kr |
| Helian Polymers, The Netherlands | PHB/PHBV | non-GMO bacteria | Unknown | helianpolymers.com |
| Biocycle, Brazil | PHB | Bacillus spp. | 100 | fapesp.br |
| Biomer, Germany | PHB | Alcaligenes latus | Unknown | biomer.de |
| Bioextrax, Sweden | PHA② | Bioextrax DSP method | Unknown | bioextrax.com |
| SABIO srl, Italy | PHA② | Organic wastes for PHA production | Unknown | www.bio-on.it |
| 1 | CHEN G Q, PATEL M K. Plastics derived from biological sources: present and future: a technical and environmental review[J]. Chemical Reviews, 2012, 112(4): 2082-2099. |
| 2 | CHEN G Q, HAJNAL I. The ‘phaome’[J]. Trends in Biotechnology, 2015, 33(10): 559-564. |
| 3 | BABU R P, O’CONNOR K, SEERAM R. Current progress on bio-based polymers and their future trends[J]. Progress in Biomaterials, 2013, 2(1): 8. |
| 4 | OLGUÍN E J, GIULIANO G, PORRO D, et al. Biotechnology for a more sustainable world[J]. Biotechnology Advances, 2012, 30(5): 931-932. |
| 5 | ANDREESSEN B, TAYLOR N, STEINBÜCHEL A. Poly(3-hydroxypropionate): a promising alternative to fossil fuel-based materials[J]. Applied and Environmental Microbiology, 2014, 80(21): 6574-6582. |
| 6 | TAN D, WANG Y, TONG Y, et al. Grand challenges for industrializing polyhydroxyalkanoates (PHAs)[J]. Trends in Biotechnology, 2021, 39(9): 953-963. |
| 7 | LEE S Y. Bacterial polyhydroxyalkanoates[J]. Biotechnology and Bioengineering, 1996, 49(1): 1-14. |
| 8 | SUDESH K, ABE H, DOI Y. Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters[J]. Progress in Polymer Science, 2000, 25(10): 1503-1555. |
| 9 | SAMUI A B, KANAI T. Polyhydroxyalkanoates based copolymers[J]. International Journal of Biological Macromolecules, 2019, 140: 522-537. |
| 10 | LI M Y, MA Y Y, ZHANG X, et al. Tailor-made polyhydroxyalkanoates by reconstructing Pseudomonas entomophila [J]. Advanced Materials, 2021, 33(41): e2102766. |
| 11 | MENG D C, SHEN R, YAO H, et al. Engineering the diversity of polyesters[J]. Current Opinion in Biotechnology, 2014, 29: 24-33. |
| 12 | CHOI S Y, CHO I J, LEE Y, et al. Microbial polyhydroxyalkanoates and nonnatural polyesters[J]. Advanced Materials, 2020, 32(35): e1907138. |
| 13 | YE J W, LIN Y N, YI X Q, et al. Synthetic biology of extremophiles: a new wave of biomanufacturing[J]. Trends in Biotechnology, 2023, 41(3): 342-357. |
| 14 | YIN J, CHEN J C, WU Q, et al. Halophiles, coming stars for industrial biotechnology[J]. Biotechnology Advances, 2015, 33(7): 1433-1442. |
| 15 | CHEN G Q. New challenges and opportunities for industrial biotechnology[J]. Microbial Cell Factories, 2012, 11: 111. |
| 16 | PARK S J, KIM T W, KIM M K, et al. Advanced bacterial polyhydroxyalkanoates: towards a versatile and sustainable platform for unnatural tailor-made polyesters[J]. Biotechnology Advances, 2012, 30(6): 1196-1206. |
| 17 | ZHENG Y, CHEN J C, MA Y M, et al. Engineering biosynthesis of polyhydroxyalkanoates (PHA) for diversity and cost reduction[J]. Metabolic Engineering, 2020, 58: 82-93. |
| 18 | AMASS W, AMASS A, TIGHE B. A review of biodegradable polymers: uses, current developments in the synthesis and characterization of biodegradable polyesters, blends of biodegradable polymers and recent advances in biodegradation studies[J]. Polymer International, 1998, 47(2): 89-144. |
| 19 | CHEN G Q. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry[J]. Chemical Society Reviews, 2009, 38(8): 2434-2446. |
| 20 | CHOI S Y, RHIE M N, KIM H T, et al. Metabolic engineering for the synthesis of polyesters: a 100-year journey from polyhydroxyalkanoates to non-natural microbial polyesters[J]. Metabolic Engineering, 2020, 58: 47-81. |
| 21 | WU Q, WANG Y, CHEN G Q. Medical application of microbial biopolyesters polyhydroxyalkanoates[J]. Artificial Cells, Blood Substitutes, and Immobilization Biotechnology, 2009, 37(1): 1-12. |
| 22 | WANG Y, BIAN Y Z, WU Q, et al. Evaluation of three-dimensional scaffolds prepared from poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) for growth of allogeneic chondrocytes for cartilage repair in rabbits[J]. Biomaterials, 2008, 29(19): 2858-2868. |
| 23 | LUO Z, WU Y L, LI Z B, et al. Recent progress in polyhydroxyalkanoates-based copolymers for biomedical applications[J]. Biotechnology Journal, 2019, 14(12): e1900283. |
| 24 | HOFFMAN A S. Stimuli-responsive polymers: Biomedical applications and challenges for clinical translation[J]. Advanced Drug Delivery Reviews, 2013, 65(1): 10-16. |
| 25 | KE Y, ZHANG X Y, RAMAKRISHNA S, et al. Reactive blends based on polyhydroxyalkanoates: preparation and biomedical application[J]. Materials Science & Engineering C, Materials for Biological Applications, 2017, 70(Pt 2): 1107-1119. |
| 26 | BIAN Y Z, WANG Y, AIBAIDOULA G, et al. Evaluation of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) conduits for peripheral nerve regeneration[J]. Biomaterials, 2009, 30(2): 217-225. |
| 27 | SODIAN R, HOERSTRUP S P, SPERLING J S, et al. Tissue engineering of heart valves: in vitro experiences[J]. Annals of Thoracic Surgery, 2000, 70(1): 140-144. |
| 28 | WEI D X, DAO J W, CHEN G Q. A micro-ark for cells: highly open porous polyhydroxyalkanoate microspheres as injectable scaffolds for tissue regeneration[J]. Advanced Materials, 2018, 30(31): e1802273. |
| 29 | ZHANG X, LI Z H, CHE X M, et al. Synthesis and characterization of polyhydroxyalkanoate organo/hydrogels[J]. Biomacromolecules, 2019, 20(9): 3303-3312. |
| 30 | FAN F, TAN D, SHANG S, et al. Poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biopolyester based nanoparticles as NVP-BEZ235 delivery vehicle for tumor targeting therapy[J]. Biomacromolecules, 2019, 20(9): 3313-3323. |
| 31 | ZHANG J Y, SHISHATSKAYA E I, VOLOVA T G, et al. Polyhydroxyalkanoates (PHA) for therapeutic applications[J]. Materials Science & Engineering C, Materials for Biological Applications, 2018, 86: 144-150. |
| 32 | ZHANG J Y, CAO Q, LI S W, et al. 3-Hydroxybutyrate methyl ester as a potential drug against Alzheimer’s disease via mitochondria protection mechanism[J]. Biomaterials, 2013, 34(30): 7552-7562. |
| 33 | LAN L H, ZHAO H, CHEN J C, et al. Engineering Halomonas spp. as a low-cost production host for production of bio-surfactant protein PhaP[J]. Biotechnology Journal, 2016, 11(12): 1595-1604. |
| 34 | WANG X, JIANG X R, WU F Q, et al. Microbial poly-3-hydroxybutyrate (PHB) as a feed additive for fishes and piglets[J]. Biotechnology Journal, 2019, 14(12): e1900132. |
| 35 | ZHANG X J, LUO R C, WANG Z, et al. Application of (R)-3-hydroxyalkanoate methyl esters derived from microbial polyhydroxyalkanoates as novel biofuels[J]. Biomacromolecules, 2009, 10(4): 707-711. |
| 36 | RYDZ J, SIKORSKA W, MUSIOŁ M, et al. 3D-printed polyester-based prototypes for cosmetic applications-future directions at the forensic engineering of advanced polymeric materials[J]. Materials, 2019, 12(6): 994. |
| 37 | DE LAS HERAS ALARCON C, PENNADAM S, ALEXANDER C. Stimuli responsive polymers for biomedical applications[J]. Chemical Society Reviews, 2005, 34(3): 276-285. |
| 38 | MA Y M, WEI D X, YAO H, et al. Synthesis, characterization and application of thermoresponsive polyhydroxyalkanoate-graft-poly(N-isopropylacrylamide)[J]. Biomacromolecules, 2016, 17(8): 2680-2690. |
| 39 | YU L P, ZHANG X, WEI D X, et al. Highly efficient fluorescent material based on rare-earth-modified polyhydroxyalkanoates[J]. Biomacromolecules, 2019, 20(9): 3233-3241. |
| 40 | LEONG Y K, SHOW P L, OOI C W, et al. Current trends in polyhydroxyalkanoates (PHAs) biosynthesis: insights from the recombinant Escherichia coli [J]. Journal of Biotechnology, 2014, 180: 52-65. |
| 41 | FERRE-GUELL A, WINTERBURN J. Biosynthesis and characterization of polyhydroxyalkanoates with controlled composition and microstructure[J]. Biomacromolecules, 2018, 19(3): 996-1005. |
| 42 | GAHLAWAT G, SRIVASTAVA A K. Model-based nutrient feeding strategies for the increased production of polyhydroxybutyrate (PHB) by Alcaligenes latus [J]. Applied Biochemistry and Biotechnology, 2017, 183(2): 530-542. |
| 43 | ARIKAWA H, MATSUMOTO K, FUJIKI T. Polyhydroxyalkanoate production from sucrose by Cupriavidus necator strains harboring csc genes from Escherichia coli W[J]. Applied Microbiology and Biotechnology, 2017, 101(20): 7497-7507. |
| 44 | RYU H W, HAHN S K, CHANG Y K, et al. Production of poly(3-hydroxybutyrate) by high cell density fed-batch culture of Alcaligenes eutrophus with phospate limitation[J]. Biotechnology and Bioengineering, 1997, 55(1): 28-32. |
| 45 | TAN D, XUE Y S, AIBAIDULA G, et al. Unsterile and continuous production of polyhydroxybutyrate by Halomonas TD01[J]. Bioresource Technology, 2011, 102(17): 8130-8136. |
| 46 | QIU Y Z, HAN J, CHEN G Q. Metabolic engineering of Aeromonas hydrophila for the enhanced production of poly (3-hydroxybutyrate-co-3-hydroxyhexanoate)[J]. Applied Microbiology and Biotechnology, 2006, 69(5): 537-542. |
| 47 | JIAN J, LI Z J, YE H M, et al. Metabolic engineering for microbial production of polyhydroxyalkanoates consisting of high 3-hydroxyhexanoate content by recombinant Aeromonas hydrophila [J]. Bioresource Technology, 2010, 101(15): 6096-6102. |
| 48 | CHUNG A L, JIN H L, HUANG L J, et al. Biosynthesis and characterization of poly(3-hydroxydodecanoate) by β-oxidation inhibited mutant of Pseudomonas entomophila L48[J]. Biomacromolecules, 2011, 12(10): 3559-3566. |
| 49 | LI S J, CAI L W, WU L P, et al. Microbial synthesis of functional homo-, random, and block polyhydroxyalkanoates by β-oxidation deleted Pseudomonas entomophila [J]. Biomacromolecules, 2014, 15(6): 2310-2319. |
| 50 | LEE S Y, WONG H H, CHOI J I, et al. Production of medium-chain-length polyhydroxyalkanoates by high-cell-density cultivation of Pseudomonas putida under phosphorus limitation[J]. Biotechnology and Bioengineering, 2000, 68(4): 466-470. |
| 51 | OUYANG S P, LIU Q, FANG L, et al. Construction of pha-operon-defined knockout mutants of Pseudomonas putida KT2442 and their applications in poly(hydroxyalkanoate) production[J]. Macromolecular Bioscience, 2007, 7(2): 227-233. |
| 52 | POBLETE-CASTRO I, BINGER D, RODRIGUES A, et al. In-silico-driven metabolic engineering of Pseudomonas putida for enhanced production of poly-hydroxyalkanoates[J]. Metabolic Engineering, 2013, 15: 113-123. |
| 53 | TRIPATHI L, WU L P, DECHUAN M, et al. Pseudomonas putida KT2442 as a platform for the biosynthesis of polyhydroxyalkanoates with adjustable monomer contents and compositions[J]. Bioresource Technology, 2013, 142: 225-231. |
| 54 | SHEN R, CAI L W, MENG D C, et al. Benzene containing polyhydroxyalkanoates homo- and copolymers synthesized by genome edited Pseudomonas entomophila [J]. Science China Life Sciences, 2014, 57(1): 4-10. |
| 55 | WANG Y, CHUNG A, CHEN G Q. Synthesis of medium-chain-length polyhydroxyalkanoate homopolymers, random copolymers, and block copolymers by an engineered strain of Pseudomonas entomophila [J]. Advanced Healthcare Materials, 2017, 6(7): 1601017. |
| 56 | CHEN Q, WANG Q, WEI G Q, et al. Production in Escherichia coli of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) with differing monomer compositions from unrelated carbon sources[J]. Applied and Environmental Microbiology, 2011, 77(14): 4886-4893. |
| 57 | HOKAMURA A, WAKIDA I, MIYAHARA Y, et al. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) by recombinant Escherichia coli from glucose[J]. Journal of Bioscience and Bioengineering, 2015, 120(3): 305-310. |
| 58 | LI T, YE J W, SHEN R, et al. Semirational approach for ultrahigh poly(3-hydroxybutyrate) accumulation in Escherichia coli by combining one-step library construction and high-throughput screening[J]. ACS Synthetic Biology, 2016, 5(11): 1308-1317. |
| 59 | WANG X, HAN J N, ZHANG X, et al. Reversible thermal regulation for bifunctional dynamic control of gene expression in Escherichia coli [J]. Nature Communications, 2021, 12(1): 1411. |
| 60 | PARK S J, AHN W S, GREEN P R, et al. Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by metabolically engineered Escherichia coli strains[J]. Biomacromolecules, 2001, 2(1): 248-254. |
| 61 | WANG F, LEE S Y. High cell density culture of metabolically engineered Escherichia coli for the production of poly(3-hydroxybutyrate) in a defined medium[J]. Biotechnology and Bioengineering, 1998, 58(2-3): 325-328. |
| 62 | YANG J E, CHOI Y J, LEE S J, et al. Metabolic engineering of Escherichia coli for biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from glucose[J]. Applied Microbiology and Biotechnology, 2014, 98(1): 95-104. |
| 63 | ZHUANG Q Q, QI Q S. Engineering the pathway in Escherichia coli for the synthesis of medium-chain-length polyhydroxyalkanoates consisting of both even- and odd-chain monomers[J]. Microbial Cell Factories, 2019, 18(1): 135. |
| 64 | CHOI J I, LEE S Y, HAN K. Cloning of the Alcaligenes latus polyhydroxyalkanoate biosynthesis genes and use of these genes for enhanced production of poly(3-hydroxybutyrate) in Escherichia coli [J]. Applied and Environmental Microbiology, 1998, 64(12): 4897-4903. |
| 65 | LEE S H, OH D H, AHN W S, et al. Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by high-cell-density cultivation of Aeromonas hydrophila [J]. Biotechnology and Bioengineering, 2000, 67(2): 240-244. |
| 66 | YE J W, HUANG W Z, WANG D S, et al. Pilot scale-up of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) production by Halomonas bluephagenesis via cell growth adapted optimization process[J]. Biotechnology Journal, 2018, 13(5): e1800074. |
| 67 | ZHUANG Q Q, WANG Q, LIANG Q F, et al. Synthesis of polyhydroxyalkanoates from glucose that contain medium-chain-length monomers via the reversed fatty acid β-oxidation cycle in Escherichia coli [J]. Metabolic Engineering, 2014, 24: 78-86. |
| 68 | MENG D C, CHEN G Q. Synthetic biology of polyhydroxyalkanoates (PHA)[J]. Advances in Biochemical Engineering/Biotechnology, 2018, 162: 147-174. |
| 69 | ZHOU Q, SHI Z Y, MENG D C, et al. Production of 3-hydroxypropionate homopolymer and poly(3-hydroxypropionate-co-4-hydroxybutyrate) copolymer by recombinant Escherichia coli [J]. Metabolic Engineering, 2011, 13(6): 777-785. |
| 70 | MENG D C, WANG Y, WU L P, et al. Production of poly (3-hydroxypropionate) and poly(3-hydroxybutyrate-co-3-hydroxypropionate) from glucose by engineering Escherichia coli [J]. Metabolic Engineering, 2015, 29: 189-195. |
| 71 | YU L P, YAN X, ZHANG X, et al. Biosynthesis of functional polyhydroxyalkanoates by engineered Halomonas bluephagenesis [J]. Metabolic Engineering, 2020, 59: 119-130. |
| 72 | YE J W, HU D K, YIN J, et al. Stimulus response-based fine-tuning of polyhydroxyalkanoate pathway in Halomonas [J]. Metabolic Engineering, 2020, 57: 85-95. |
| 73 | MA H, ZHAO Y Q, HUANG W Z, et al. Rational flux-tuning of Halomonas bluephagenesis for co-production of bioplastic PHB and ectoine[J]. Nature Communications, 2020, 11(1): 3313. |
| 74 | YE J W, HU D K, CHE X M, et al. Engineering of Halomonas bluephagenesis for low cost production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) from glucose[J]. Metabolic Engineering, 2018, 47: 143-152. |
| 75 | YAN X, LIU X, YU L P, et al. Biosynthesis of diverse α,ω-diol-derived polyhydroxyalkanoates by engineered Halomonas bluephagenesis [J]. Metabolic Engineering, 2022, 72: 275-288. |
| 76 | HAN J, WU L P, HOU J, et al. Biosynthesis, characterization, and hemostasis potential of tailor-made poly(3-hydroxybutyrate-co-3-hydroxyvalerate) produced by Haloferax mediterranei [J]. Biomacromolecules, 2015, 16(2): 578-588. |
| 77 | HAN J, WU L P, LIU X B, et al. Biodegradation and biocompatibility of haloarchaea-produced poly (3-hydroxybutyrate-co-3-hydroxyvalerate) copolymers[J]. Biomaterials, 2017, 139: 172-186. |
| 78 | QUILLAGUAMÁN J, GUZMÁN H, VAN-THUOC D, et al. Synthesis and production of polyhydroxyalkanoates by halophiles: current potential and future prospects[J]. Applied Microbiology and Biotechnology, 2010, 85(6): 1687-1696. |
| 79 | QUILLAGUAMÁN J, DELGADO O, MATTIASSON B, et al. Poly(β-hydroxybutyrate) production by a moderate halophile, Halomonas boliviensis LC1[J]. Enzyme and Microbial Technology, 2006, 38(1/2): 148-154. |
| 80 | CHEN G Q, JIANG X R. Next generation industrial biotechnology based on extremophilic bacteria[J]. Current Opinion in Biotechnology, 2018, 50: 94-100. |
| 81 | YU L P, WU F Q, CHEN G Q. Next-generation industrial biotechnology-transforming the current industrial biotechnology into competitive processes[J]. Biotechnology Journal, 2019, 14(9): e1800437. |
| 82 | HAJNAL I, CHEN X B, CHEN G Q. A novel cell autolysis system for cost-competitive downstream processing[J]. Applied Microbiology and Biotechnology, 2016, 100(21): 9103-9110. |
| 83 | CHEN G Q, ZHANG X, LIU X, et al. Halomonas spp., as chassis for low-cost production of chemicals[J]. Applied Microbiology and Biotechnology, 2022, 106(21): 6977-6992. |
| 84 | JIANG X R, CHEN G Q. Morphology engineering of bacteria for bio-production[J]. Biotechnology Advances, 2016, 34(4): 435-440. |
| 85 | WU H, CHEN J C, CHEN G Q. Engineering the growth pattern and cell morphology for enhanced PHB production by Escherichia coli [J]. Applied Microbiology and Biotechnology, 2016, 100(23): 9907-9916. |
| 86 | JIANG X R, WANG H, SHEN R, et al. Engineering the bacterial shapes for enhanced inclusion bodies accumulation[J]. Metabolic Engineering, 2015, 29: 227-237. |
| 87 | WANG Y, WU H, JIANG X R, et al. Engineering Escherichia coli for enhanced production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) in larger cellular space[J]. Metabolic Engineering, 2014, 25: 183-193. |
| 88 | ELHADI D, LV L, JIANG X R, et al. CRISPRi engineering E. coli for morphology diversification[J]. Metabolic Engineering, 2016, 38: 358-369. |
| 89 | SHEN R, NING Z Y, LAN Y X, et al. Manipulation of polyhydroxyalkanoate granular sizes in Halomonas bluephagenesis [J]. Metabolic Engineering, 2019, 54: 117-126. |
| 90 | YUE H T, LING C, YANG T, et al. A seawater-based open and continuous process for polyhydroxyalkanoates production by recombinant Halomonas campaniensis LS21 grown in mixed substrates[J]. Biotechnology for Biofuels, 2014, 7(1): 108. |
| 91 | SILVA-ROCHA R, MARTÍNEZ-GARCÍA E, CALLES B, et al. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes[J]. Nucleic Acids Research, 2013, 41(Database issue): D666-D675. |
| 92 | TAN D, WU Q, CHEN J C, et al. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates[J]. Metabolic Engineering, 2014, 26: 34-47. |
| 93 | FU X Z, TAN D, AIBAIDULA G, et al. Development of Halomonas TD01 as a host for open production of chemicals[J]. Metabolic Engineering, 2014, 23: 78-91. |
| 94 | XU T, MITRA R, TAN D, et al. Utilization of gene manipulation system for advancing the biotechnological potential of halophiles: a review[J]. Biotechnology Advances, 2024, 70: 108302. |
| 95 | MA Y Y, YE J W, LIN Y N, et al. Flux optimization using multiple promoters in Halomonas bluephagenesis as a model chassis of the next generation industrial biotechnology[J]. Metabolic Engineering, 2024, 81: 249-261. |
| 96 | XU T, CHEN J Y, MITRA R, et al. Deficiency of exopolysaccharides and O-antigen makes Halomonas bluephagenesis self-flocculating and amenable to electrotransformation[J]. Communications Biology, 2022, 5(1): 623. |
| 97 | WANG Z Y, QIN Q, ZHENG Y F, et al. Engineering the permeability of Halomonas bluephagenesis enhanced its chassis properties[J]. Metabolic Engineering, 2021, 67: 53-66. |
| 98 | QIN Q, LING C, ZHAO Y Q, et al. CRISPR/Cas9 editing genome of extremophile Halomonas spp.[J]. Metabolic Engineering, 2018, 47: 219-229. |
| 99 | LIU C Y, YUE Y X, XUE Y F, et al. CRISPR-Cas9 assisted non-homologous end joining genome editing system of Halomonas bluephagenesis for large DNA fragment deletion[J]. Microbial Cell Factories, 2023, 22(1): 211. |
| 100 | SHEN R, YIN J, YE J W, et al. Promoter engineering for enhanced P(3HB-co-4HB) production by Halomonas bluephagenesis [J]. ACS Synthetic Biology, 2018, 7(8): 1897-1906. |
| 101 | ZHAO H, ZHANG H Q M, CHEN X B, et al. Novel T7-like expression systems used for Halomonas [J]. Metabolic Engineering, 2017, 39: 128-140. |
| 102 | WANG L J, JIANG X R, HOU J, et al. Engineering Halomonas bluephagenesis via small regulatory RNAs[J]. Metabolic Engineering, 2022, 73: 58-69. |
| 103 | XU M M, CHANG Y, ZHANG Y Y, et al. Development and application of transcription terminators for polyhydroxylkanoates production in halophilic Halomonas bluephagenesis TD01[J]. Frontiers in Microbiology, 2022, 13: 941306. |
| 104 | MA Y Y, ZHENG X R, LIN Y N, et al. Engineering an oleic acid-induced system for Halomonas, E. coli and Pseudomonas [J]. Metabolic Engineering, 2022, 72: 325-336. |
| 105 | CHEN Y, CHEN X Y, DU H T, et al. Chromosome engineering of the TCA cycle in Halomonas bluephagenesis for production of copolymers of 3-hydroxybutyrate and 3-hydroxyvalerate (PHBV)[J]. Metabolic Engineering, 2019, 54: 69-82. |
| 106 | REN Y L, LING C, HAJNAL I, et al. Construction of Halomonas bluephagenesis capable of high cell density growth for efficient PHA production[J]. Applied Microbiology and Biotechnology, 2018, 102(10): 4499-4510. |
| 107 | OUYANG P F, WANG H, HAJNAL I, et al. Increasing oxygen availability for improving poly(3-hydroxybutyrate) production by Halomonas [J]. Metabolic Engineering, 2018, 45: 20-31. |
| 108 | LING C, QIAO G Q, SHUAI B W, et al. Engineering NADH/NAD+ ratio in Halomonas bluephagenesis for enhanced production of polyhydroxyalkanoates (PHA)[J]. Metabolic Engineering, 2018, 49: 275-286. |
| 109 | LI T, CHEN X B, CHEN J C, et al. Open and continuous fermentation: products, conditions and bioprocess economy[J]. Biotechnology Journal, 2014, 9(12): 1503-1511. |
| 110 | SHAHZAD K, NARODOSLAWSKY M, SAGIR M, et al. Techno-economic feasibility of waste biorefinery: using slaughtering waste streams as starting material for biopolyester production[J]. Waste Management, 2017, 67: 73-85. |
| 111 | TARRAHI R, FATHI Z, SEYDIBEYOĞLU M Ö, et al. Polyhydroxyalkanoates (PHA): from production to nanoarchitecture[J]. International Journal of Biological Macromolecules, 2020, 146: 596-619. |
| 112 | ALI M, BROCCHINI S. Synthetic approaches to uniform polymers[J]. Advanced Drug Delivery Reviews, 2006, 58(15): 1671-1687. |
| 113 | AYNSLEY M, HOFLAND A, MORRIS A J, et al. Artificial intelligence and the supervision of bioprocesses (real-time knowledge-based systems and neural networks)[J]. Advances in Biochemical Engineering/Biotechnology, 1993, 48: 1-27. |
| 114 | GUERRA A, VON STOSCH M, GLASSEY J. Toward biotherapeutic product real-time quality monitoring[J]. Critical Reviews in Biotechnology, 2019, 39(3): 289-305. |
| [1] | LIU Xiaoyue, WANG Pandi, WU Gang, LIU Fang. Efficient biosynthesis of glucoraphanin in Brassicaceae crops by genetic engineering [J]. Synthetic Biology Journal, 2025, 6(1): 136-156. |
| [2] | WEN Yanhua, LIU Hedong, CAO Chunlai, WU Ruibo. Applications of protein engineering in pharmaceutical industry [J]. Synthetic Biology Journal, 2025, 6(1): 65-86. |
| [3] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [4] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| [5] | WANG Kai, ZHANG Wan, HUANG Yunhai, ZHANG Lixin, LOU Chunbo. Application of phage therapy in the treatment of intracellular pathogens [J]. Synthetic Biology Journal, 2023, 4(4): 676-689. |
| [6] | TANG Yiming, YAO Yifei, YANG Zhongyuan, ZHOU Yun, WANG Zichao, WEI Guanghong. Pathological aggregation and liquid-liquid phase separation of proteins associated with neurodegenerative diseases [J]. Synthetic Biology Journal, 2023, 4(3): 590-610. |
| [7] | CHEN Qingli, TONG Yigang. Merging the frontiers: synthetic biology for advanced bacteriophage design [J]. Synthetic Biology Journal, 2023, 4(2): 283-300. |
| [8] | Zhaoying YANG, Fan ZHANG, Jianwen GUO, Weiping GAO. Biosynthesis of elastin-like polypeptides and their applications in drug delivery [J]. Synthetic Biology Journal, 2022, 3(4): 728-747. |
| [9] | Qian WANG, Qingsheng QI. Low-carbon biomanufacturing of polyhydroxyalkanoates: analysis and application based on carbon conversion rate [J]. Synthetic Biology Journal, 2022, 3(4): 748-762. |
| [10] | Xiaosheng LIANG, Yongchao GUO, Dong MEN, Xian’en ZHANG. Hybrid systems of virus and nano-gold conducting networks for electrochemical analysis [J]. Synthetic Biology Journal, 2022, 3(2): 415-427. |
| [11] | Jiaoyu JIN, Jiahai ZHOU. The mystery of Z-genome biosynthesis has been elucidated [J]. Synthetic Biology Journal, 2022, 3(1): 1-5. |
| [12] | Jianming LYU, Huan ZHAO, Dan HU, Hao GAO. Biosynthesis of alkyne moiety in natural products and application of alkyne biosynthetic machineries [J]. Synthetic Biology Journal, 2021, 2(5): 734-750. |
| [13] | Zhiguo SU. Great impact of Professor Daniel I.C. Wang and BPEC on development of biochemical engineering [J]. Synthetic Biology Journal, 2021, 2(4): 470-481. |
| [14] | Hui ZHANG, Yaomeng YUAN, Chong ZHANG, Song YANG, Xinhui XING. Research progresses and future prospects of synthetic methylotrophic cell factory for methanol assimilation [J]. Synthetic Biology Journal, 2021, 2(2): 222-233. |
| [15] | Shengjian YUAN, Yingfei MA. Advances and applications of phage synthetic biology [J]. Synthetic Biology Journal, 2020, 1(6): 635-655. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||