Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (6): 1250-1261.DOI: 10.12211/2096-8280.2022-024
• Invited Review • Previous Articles Next Articles
Designing, building and rapid prototyping of biosynthesis module based on cell-free system
TANG Shiming, HU Jiyuan, ZHENG Suiping, HAN Shuangyan, LIN Ying
- School of Biology and Biological Engineering,South China University of Technology,Guangzhou 510006,Guangdong,China
-
Received:2022-04-21Revised:2022-09-06Online:2023-01-17Published:2022-12-31 -
Contact:LIN Ying
基于无细胞体系的生物合成代谢模块设计、构建与快速途径原型
唐士茗, 胡纪元, 郑穗平, 韩双艳, 林影
- 华南理工大学,生物科学与工程学院,广东 广州 510006
-
通讯作者:林影 -
作者简介:唐士茗 (1995—),男,博士研究生。研究方向为酶工程、合成生物学。E-mail:202010108586@mail.scut.edu.cn林影 (1962—),女,博士,教授。研究方向为酶学与酶工程、合成生物学等,国家重点研发计划“合成生物学”专项项目负责人。E-mail:feylin@scut.edu.cn -
基金资助:国家重点研发计划(2018YFA0901700)
CLC Number:
Cite this article
TANG Shiming, HU Jiyuan, ZHENG Suiping, HAN Shuangyan, LIN Ying. Designing, building and rapid prototyping of biosynthesis module based on cell-free system[J]. Synthetic Biology Journal, 2022, 3(6): 1250-1261.
唐士茗, 胡纪元, 郑穗平, 韩双艳, 林影. 基于无细胞体系的生物合成代谢模块设计、构建与快速途径原型[J]. 合成生物学, 2022, 3(6): 1250-1261.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-024
| 衡量标准 | 细胞生物合成 | 无细胞生物合成 |
|---|---|---|
| 途径设计 | 胞内代谢网络复杂 | 自由灵活 |
| 生产效率 | 碳通量被用于细胞生长及产生副产物 | 碳通量最大程度流向目标化合物 |
| 物质传输 | 具有选择性屏障 | 直接添加底物,产物易提取 |
| 产物分离纯化 | 副产物较多 | 较容易 |
| 毒性影响 | 毒性物质影响细胞生长 | 没有生存限制 |
| 成本 | 较低 | 酶的制备和辅因子成本 |
| 成熟性 | 多年实践经验 | 初具规模 |
Tab. 1 Comparison of cellular and cell-free biosynthesis
| 衡量标准 | 细胞生物合成 | 无细胞生物合成 |
|---|---|---|
| 途径设计 | 胞内代谢网络复杂 | 自由灵活 |
| 生产效率 | 碳通量被用于细胞生长及产生副产物 | 碳通量最大程度流向目标化合物 |
| 物质传输 | 具有选择性屏障 | 直接添加底物,产物易提取 |
| 产物分离纯化 | 副产物较多 | 较容易 |
| 毒性影响 | 毒性物质影响细胞生长 | 没有生存限制 |
| 成本 | 较低 | 酶的制备和辅因子成本 |
| 成熟性 | 多年实践经验 | 初具规模 |
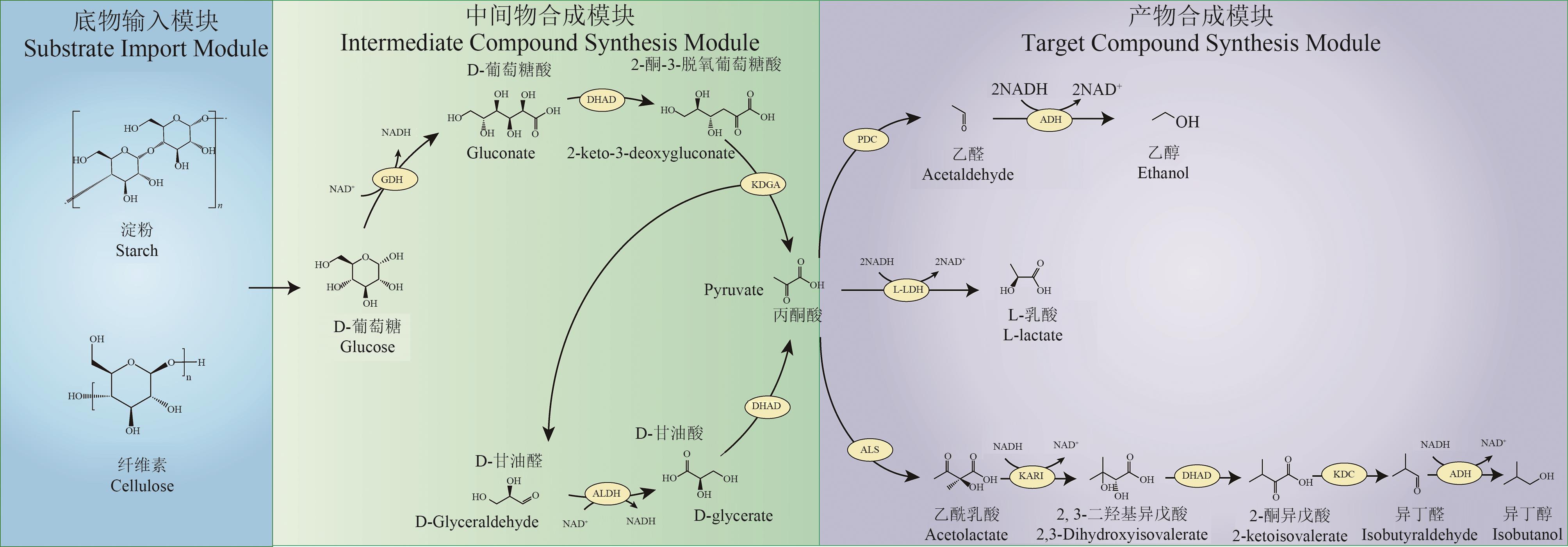
Fig. 1 Modular design of cell-free biosynthetic pathway from starch-base materialsGDH—glucose dehydrogenase; DHAD—dihydroxy acid dehydratase; KDGA—2-keto-3-desoxy gluconate aldolase; ALDH—glyceraldehyde dehydrogenase; L-LDH—L-lactate dehydrogenase; PDC—pyruvate decarboxylase; ADH—alcohol dehydrogenase; ALS—acetolactate synthase; KARI—ketolacid reductoisomerase; KDC—2-ketoacid decarboxylase
| 1 | KORMAN T P, OPGENORTH P H, BOWIE J U. A synthetic biochemistry platform for cell free production of monoterpenes from glucose[J]. Nature Communications, 2017, 8: 15526. |
| 2 | KRUTSAKORN B, HONDA K, YE X T, et al. In vitro production of n-butanol from glucose[J]. Metabolic Engineering, 2013, 20: 84-91. |
| 3 | LU X Y, LIU Y W, YANG Y Q, et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design[J]. Nature Communications, 2019, 10: 1378. |
| 4 | CAI T, SUN H B, QIAO J, et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide[J]. Science, 2021, 373(6562): 1523-1527. |
| 5 | CHEN R B, GAO J Q, YU W, et al. Engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast[J]. Nature Chemical Biology, 2022, 18(5): 520-529. |
| 6 | XU Y M, WANG X L, ZHANG C Y, et al. De novo biosynthesis of rubusoside and rebaudiosides in engineered yeasts[J]. Nature Communications, 2022, 13: 3040. |
| 7 | FANG H, LI D, KANG J, et al. Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12 [J]. Nature Communications, 2018, 9: 4917. |
| 8 | KIM H M, CHAE T U, CHOI S Y, et al. Engineering of an oleaginous bacterium for the production of fatty acids and fuels[J]. Nature Chemical Biology, 2019, 15(7): 721-729. |
| 9 | LU Y. Cell-free synthetic biology: Engineering in an open world[J]. Synthetic and Systems Biotechnology, 2017, 2(1): 23-27. |
| 10 | GRUBBE W S, RASOR B J, KRÜGER A, et al. Cell-free styrene biosynthesis at high titers[J]. Metabolic Engineering, 2020, 61: 89-95. |
| 11 | TANG S M, LIAO D C, LI X W, et al. Cell-free biosynthesis system: Methodology and perspective of in vitro efficient platform for pyruvate biosynthesis and transformation[J]. ACS Synthetic Biology, 2021, 10(10): 2417-2433. |
| 12 | YI T, LIM H J, LEE S J, et al. Synthesis of (R, R)-2, 3-butanediol from starch in a hybrid cell-free reaction system[J]. Journal of Industrial and Engineering Chemistry, 2018, 67: 231-235. |
| 13 | HONDA K, KIMURA K, NINH P H, et al. In vitro bioconversion of chitin to pyruvate with thermophilic enzymes[J]. Journal of Bioscience and Bioengineering, 2017, 124(3): 296-301. |
| 14 | LIU Z, ZHANG Y C, JIA X G, et al. In vitro reconstitution and optimization of the entire pathway to convert glucose into fatty acid[J]. ACS Synthetic Biology, 2017, 6(4): 701-709. |
| 15 | LI Y J, SHI T, HAN P P, et al. Thermodynamics-driven production of value-added d-allulose from inexpensive starch by an in vitro enzymatic synthetic biosystem[J]. ACS Catalysis, 2021, 11(9): 5088-5099. |
| 16 | WEI X L, LI Q Z, HU C C, et al. An ATP-free in vitro synthetic enzymatic biosystem facilitating one-pot stoichiometric conversion of starch to mannitol[J]. Applied Microbiology and Biotechnology, 2021, 105(5): 1913-1924. |
| 17 | ZHONG C, YOU C, WEI P, et al. Thermal cycling cascade biocatalysis of myo-inositol synthesis from sucrose[J]. ACS Catalysis, 2017, 7(9): 5992-5999. |
| 18 | SINGH R K, SINGH R, SIVAKUMAR D, et al. Insights into cell-free conversion of CO2 to chemicals by a multienzyme cascade reaction[J]. ACS Catalysis, 2018, 8(12): 11085-11093. |
| 19 | BEER B, PICK A, SIEBER V. In vitro metabolic engineering for the production of α-ketoglutarate[J]. Metabolic Engineering, 2017, 40: 5-13. |
| 20 | LI C, ZHANG Z D, WANG J. A Thermophilic biofunctional multienzyme cascade reaction for cell-rree synthesis of salvianic acid A and 3,4-dihydroxymandelic acid [J]. ACS Sustainable Chemistry & Engineering, 2019, 7(22): 18247-18253. |
| 21 | SUN S S, WEI X L, YOU C. The construction of an in vitro synthetic enzymatic biosystem that facilitates laminaribiose biosynthesis from maltodextrin and glucose[J]. Biotechnology Journal, 2019, 14(4): 1800493. |
| 22 | KIM T H, KANG S H, HAN J E, et al. Multilayer engineering of enzyme cascade catalysis for one-pot preparation of nylon monomers from renewable fatty acids [J]. ACS Catalysis, 2020, 10(9): 4871-4878. |
| 23 | OPGENORTH P H, KORMAN T P, BOWIE J U. A synthetic biochemistry module for production of bio-based chemicals from glucose[J]. Nature Chemical Biology, 2016, 12(6): 393-395. |
| 24 | WANG W, YANG J G, SUN Y X, et al. Artificial ATP-free in vitro synthetic enzymatic biosystems facilitate aldolase-mediated C—C bond formation for biomanufacturing[J]. ACS Catalysis, 2020, 10(2): 1264-1271. |
| 25 | BAI X, MENG D, WEI X, et al. Facile synthesis of (-)-vibo-quercitol from maltodextrin via an in vitro synthetic enzymatic biosystem[J]. Biotechnology and Bioengineering, 2019, 116(10): 2710-2719. |
| 26 | DUDLEY Q M, ANDERSON K C, JEWETT M C. Cell-free mixing of escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis[J]. ACS Synthetic Biology, 2016, 5(12): 1578-1588. |
| 27 | LUO Y Z, LEE J K, ZHAO H M. Challenges and opportunities in synthetic biology for chemical engineers[J]. Chemical Engineering Science, 2013, 103: 115-119. |
| 28 | TANIGUCHI H, OKANO K, HONDA K. Modules for in vitro metabolic engineering: pathway assembly for bio-based production of value-added chemicals[J]. Synthetic and Systems Biotechnology, 2017, 2(2): 65-74. |
| 29 | XU P, GU Q, WANG W Y, et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli [J]. Nature Communications, 2013, 4: 1409. |
| 30 | GUTERL J K, GARBE D, CARSTEN J, et al. Cell-free metabolic engineering: production of chemicals by minimized reaction cascades[J]. ChemSusChem, 2012, 5(11): 2165-2172. |
| 31 | XIE L P, WEI X L, ZHOU X G, et al. Conversion of d-glucose to l-lactate via pyruvate by an optimized cell-free enzymatic biosystem containing minimized reactions[J]. Synthetic and Systems Biotechnology, 2018, 3(3): 204-210. |
| 32 | DUDLEY Q M, NASH C J, JEWETT M C. Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates[J]. Synthetic Biology, 2019, 4(1): ysz003. |
| 33 | DU Z Q, LIU Z Y, TAN Y S, et al. Lacto-N-biose synthesis via a modular enzymatic cascade with ATP regeneration[J]. iScience, 2021, 24(3): 102236. |
| 34 | OPGENORTH P H, KORMAN T P, BOWIE J U. A synthetic biochemistry molecular purge valve module that maintains redox balance[J]. Nature Communications, 2014, 5: 4113. |
| 35 | GMELCH T J, SPERL J M, SIEBER V. Optimization of a reduced enzymatic reaction cascade for the production of L-alanine[J]. Scientific Reports, 2019, 9: 11754. |
| 36 | WANG L, DASH S, NG C Y, et al. A review of computational tools for design and reconstruction of metabolic pathways[J]. Synthetic and Systems Biotechnology, 2017, 2(4): 243-252. |
| 37 | RAHMAN S A, ADVANI P, SCHUNK R, et al. Metabolic pathway analysis web service (Pathway Hunter Tool at CUBIC)[J]. Bioinformatics, 2005, 21(7): 1189-1193. |
| 38 | CHOU C H, CHANG W C, CHIU C M, et al. FMM: a web server for metabolic pathway reconstruction and comparative analysis[J]. Nucleic Acids Research, 2009, 37(): W129-W134. |
| 39 | DELÉPINE B, DUIGOU T, CARBONELL P, et al. RetroPath2.0: a retrosynthesis workflow for metabolic engineers[J]. Metabolic Engineering, 2018, 45: 158-170. |
| 40 | CARBONELL P, PARUTTO P, HERISSON J, et al. XTMS: pathway design in an eXTended metabolic space[J]. Nucleic Acids Research, 2014, 42(W1): W389-W394. |
| 41 | YANG X, YUAN Q Q, LUO H, et al. Systematic design and in vitro validation of novel one-carbon assimilation pathways[J]. Metabolic Engineering, 2019, 56: 142-153. |
| 42 | SEGLER M H S, PREUSS M, WALLER M P. Planning chemical syntheses with deep neural networks and symbolic AI[J]. Nature, 2018, 555(7698): 604-610. |
| 43 | LI Y Q, FANG J W. PROTS-RF: a robust model for predicting mutation-induced protein stability changes[J]. PLoS One, 2012, 7(10): e47247. |
| 44 | YANG Y, UROLAGIN S, NIROULA A, et al. PON-tstab: Protein variant stability predictor. importance of training data quality[J]. International Journal of Molecular Sciences, 2018, 19(4): 1009. |
| 45 | JOKINEN E, HEINONEN M, LÄHDESMÄKI H. mGPfusion: predicting protein stability changes with Gaussian process kernel learning and data fusion[J]. Bioinformatics, 2018, 34(13): i274-i283. |
| 46 | GIOLLO M, MARTIN A J M, WALSH I, et al. NeEMO: a method using residue interaction networks to improve prediction of protein stability upon mutation[J]. BMC Genomics, 2014, 15(): S7. |
| 47 | LI G Y, QIN Y C, FONTAINE N T, et al. Machine learning enables selection of epistatic enzyme mutants for stability against unfolding and detrimental aggregation[J]. ChemBioChem, 2021, 22(5): 904-914. |
| 48 | WANG X F, GAO P, LIU Y F, et al. Predicting thermophilic proteins by machine learning[J]. Current Bioinformatics, 2020, 15(5): 493-502. |
| 49 | WU Z, KAN S B J, LEWIS R D, et al. Machine learning-assisted directed protein evolution with combinatorial libraries[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(18): 8852-8858. |
| 50 | FOX R J, DAVIS S C, MUNDORFF E C, et al. Improving catalytic function by ProSAR-driven enzyme evolution[J]. Nature Biotechnology, 2007, 25(3): 338-344. |
| 51 | KHURANA S, RAWI R, KUNJI K, et al. DeepSol: A deep learning framework for sequence-based protein solubility prediction[J]. Bioinformatics, 2018, 34(15): 2605-2613. |
| 52 | LI Y, WANG S, UMAROV R, et al. DEEPre: sequence-based enzyme EC number prediction by deep learning[J]. Bioinformatics, 2018, 34(5): 760-769. |
| 53 | KARIM A S, DUDLEY Q M, JUMINAGA A, et al. In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design[J]. Nature Chemical Biology, 2020, 16(8): 912-919. |
| 54 | NOOR E, BAR-EVEN A, FLAMHOLZ A, et al. Pathway thermodynamics highlights kinetic obstacles in central metabolism[J]. PLoS Computational Biology, 2014, 10(2): e1003483. |
| 55 | FLAMHOLZ A, NOOR E, BAR-EVEN A, et al. eQuilibrator-the biochemical thermodynamics calculator[J]. Nucleic Acids Research, 2012, 40(D1): D770-D775. |
| 56 | WHITTAKER J W. Cell-free protein synthesis: the state of the art[J]. Biotechnology Letters, 2013, 35(2): 143-152. |
| 57 | CARLSON E D, GAN R, HODGMAN C E, et al. Cell-free protein synthesis: applications come of age[J]. Biotechnology Advances, 2012, 30(5): 1185-1194. |
| 58 | GREGORIO N E, LEVINE M Z, OZA J P. A user's guide to cell-free protein synthesis[J]. Methods and Protocols, 2019, 2(1): 24. |
| 59 | SILVERMAN A D, KARIM A S, JEWETT M C. Cell-free gene expression: an expanded repertoire of applications[J]. Nature Reviews Genetics, 2020, 21(3): 151-170. |
| 60 | SCHINN S M, BROADBENT A, BRADLEY W T, et al. Protein synthesis directly from PCR: progress and applications of cell-free protein synthesis with linear DNA[J]. New Biotechnology, 2016, 33(4): 480-487. |
| 61 | SUN Z Z, YEUNG E, HAYES C A, et al. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system[J]. ACS Synthetic Biology, 2014, 3(6): 387-397. |
| 62 | JIANG L H, ZHAO J R, LIAN J Z, et al. Cell-free protein synthesis enabled rapid prototyping for metabolic engineering and synthetic biology[J]. Synthetic and Systems Biotechnology, 2018, 3(2): 90-96. |
| 63 | GAN R, JEWETT M C. A combined cell-free transcription-translation system from Saccharomyces cerevisiae for rapid and robust protein synthe[J]. Biotechnology Journal, 2014, 9(5): 641-651. |
| 64 | HODGMAN C E, JEWETT M C. Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis[J]. Biotechnology and Bioengineering, 2013, 110(10): 2643-2654. |
| 65 | XU H L, LIU W Q, LI J. Translation related factors improve the productivity of a streptomyces-based cell-free protein synthesis system[J]. ACS Synthetic Biology, 2020, 9(5): 1221-1224. |
| 66 | TABUCHI T, YOKOBAYASHI Y. Cell-free riboswitches [J]. RSC Chemical Biology, 2021, 2(5): 1430-1440. |
| 67 | KARIM A S, JEWETT M C. Cell-free synthetic biology for pathway prototyping[J]. Methods in Enzymology, 2018, 608: 31-57. |
| 68 | KORMAN T P, SAHACHARTSIRI B, LI D, et al. A synthetic biochemistry system for the in vitro production of isoprene from glycolysis intermediates[J]. Protein Science, 2014, 23(5): 576-585. |
| 69 | ZHU F Y, ZHONG X F, HU M Z, et al. In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli [J]. Biotechnology and Bioengineering, 2014, 111(7): 1396-1405. |
| 70 | CHEN X X, ZHANG C Q, ZOU R Y, et al. Statistical experimental design guided optimization of a one-pot biphasic multienzyme total synthesis of amorpha-4, 11-diene[J]. PLoS One, 2013, 8(11): e79650. |
| 71 | RODRIGUEZ S B, LEYH T S. An enzymatic platform for the synthesis of isoprenoid precursors[J]. PLoS One, 2014, 9(8): e105594. |
| 72 | TOOGOOD H S, CHEALLAIGH A N, TAIT S, et al. Enzymatic menthol production: one-pot approach using engineered Escherichia coli [J]. ACS Synthetic Biology, 2015, 4(10): 1112-1123. |
| 73 | SIEBELS I, NOWAK S, HEIL C S, et al. Cell-free synthesis of natural compounds from genomic DNA of biosynthetic gene clusters[J]. ACS Synthetic Biology, 2020, 9(9): 2418-2426. |
| 74 | KARIM A S, JEWETT M C. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery[J]. Metabolic Engineering, 2016, 36: 116-126. |
| 75 | KAY J E, JEWETT M C. Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2, 3-butanediol[J]. Metabolic Engineering, 2015, 32: 133-142. |
| 76 | BUJARA M, PANKE S. In silico assessment of cell-free systems[J]. Biotechnology and Bioengineering, 2012, 109(10): 2620-2629. |
| 77 | DUDLEY Q M, KARIM A S, NASH C J, et al. In vitro prototyping of limonene biosynthesis using cell-free protein synthesis[J]. Metabolic Engineering, 2020, 61: 251-260. |
| 78 | 林影, 袁清焱, 梁书利. 定量检测无细胞蛋白质合成系统目的蛋白产量及筛选高催化活性酶蛋白的方法. CN114019169A [P].2022-02-08. |
| LIN Y, YUAN Q Y, LIANG S L. A method of quantification of protein production in cell-free protein synthesis and screening of enzymes with high catalytic activity, CN114019169A [P]. 2022-02-08. |
| [1] | LIU Wanqiu, JI Xiangyang, XU Huiling, LU Yicong, LI Jian. Cell-free protein synthesis system enables rapid and efficient biosynthesis of restriction endonucleases [J]. Synthetic Biology Journal, 2023, 4(4): 840-851. |
| [2] | ZENG Tao, WU Ruibo. Data-driven prediction and design for enzymatic reactions [J]. Synthetic Biology Journal, 2023, 4(3): 535-550. |
| [3] | Jiaqi HOU, Nan JIANG, Lianju MA, Yuan LU. Cell-free protein synthesis: from basic research to engineering applications [J]. Synthetic Biology Journal, 2022, 3(3): 465-486. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||