Progress in Biological Entity-Material Hybrid System for Low-Carbon Biosynthesis
Xinyi GUO1, Shuqi GUO1,2, Shuwei LI1, Ziyue JIAO1, Qiang FEI1,2
- 1.School of Chemical Engineering and Technology,Xi’an Jiaotong University,Xi’an 710049,Shaanxi,China
2.Xi’an Key Laboratory of C1 Compound Bioconversion Technology,Xi’an 710049,Shaanxi,China
-
Received:2024-09-18Revised:2024-12-26Published:2024-12-28 -
Contact:Qiang FEI
基于生物体-材料杂化体系的低碳生物合成的研究进展
郭心怡1, 郭树奇1,2, 李曙伟1, 焦子悦1, 费强1,2
- 1.西安交通大学,化学工程与技术学院,陕西 西安 710049
2.西安市一碳化合物生物转化技术重点实验室,陕西 西安 710049
-
通讯作者:费强 -
作者简介:郭心怡 (1999—),女,在读博士研究生。研究方向为嗜甲烷菌细胞工厂构建及微生物-材料杂化体系转化甲烷合成化学品。E-mail:gxy9933@stu.xjtu.edu.cn费 强 (1980—),男,西安交通大学教授,博士生导师,陕西省杰青基金获得者、陕西高校青年创新团队负责人,一碳化合物生物转化技术西安市重点实验室主任,中国生物工程学会一碳生物技术专委会秘书长。近年承担国家重点研发计划、国家自然科学基金、陕西省重点研发计划等多项科研项目。以一碳气体的微生物固定及其高值化利用为研究目标,利用合成生物学和高密度发酵等技术改造和优化细胞工厂,实现食品、材料、化学品、能源等产品的生物制造。E-mail:feiqiang@xjtu.edu.cn -
基金资助:国家重点研发计划(2021YFC2103500);国家自然科学基金(2217281)
CLC Number:
Cite this article
Xinyi GUO, Shuqi GUO, Shuwei LI, Ziyue JIAO, Qiang FEI. Progress in Biological Entity-Material Hybrid System for Low-Carbon Biosynthesis[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2024-073.
郭心怡, 郭树奇, 李曙伟, 焦子悦, 费强. 基于生物体-材料杂化体系的低碳生物合成的研究进展[J]. 合成生物学, DOI: 10.12211/2096-8280.2024-073.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-073
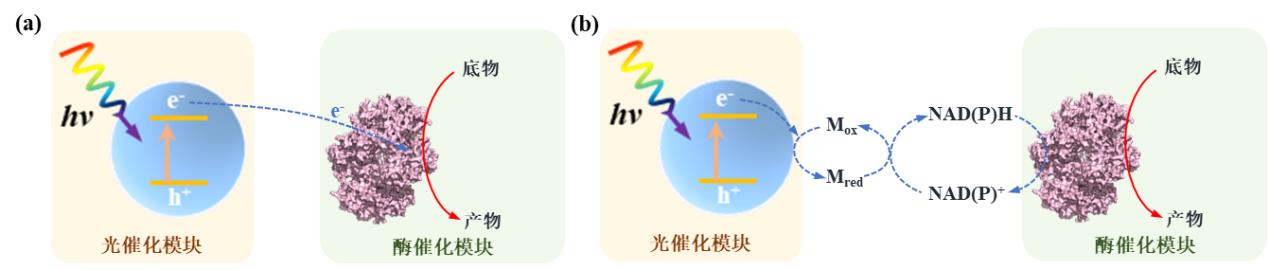
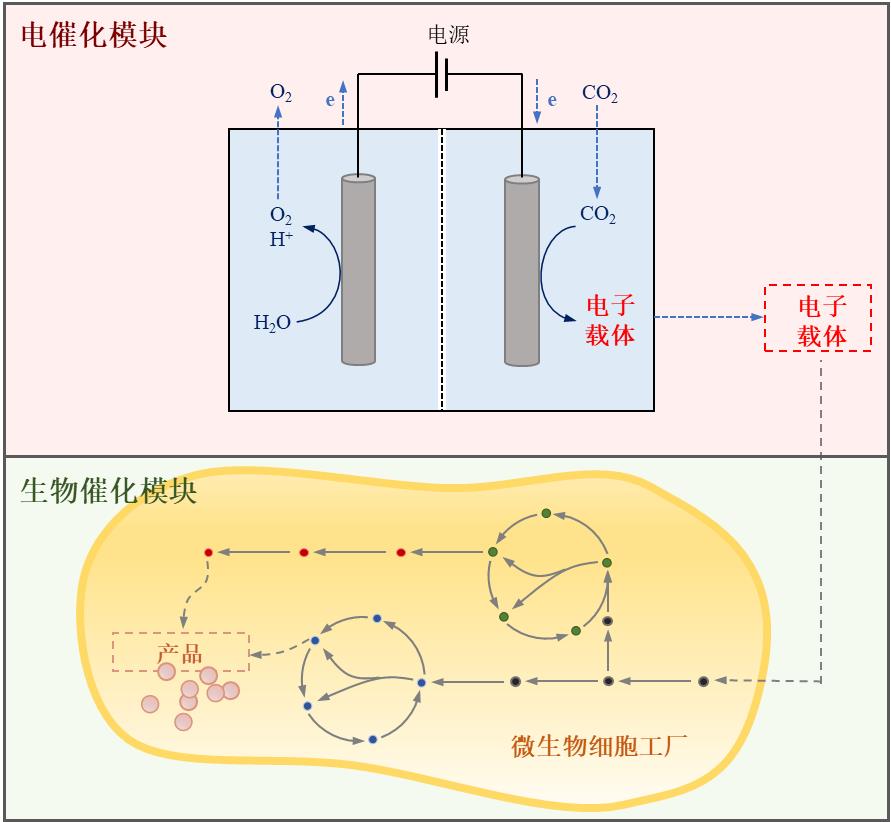
Fig. 1 Electron transport mechanism in light-driven material-enzyme hybrid system. (a) Direct electron transfer between cell and electrode; (b) Indirect electron transfer between cell and electrode. (Mox: oxidized mediator, Mred: reduced mediator)
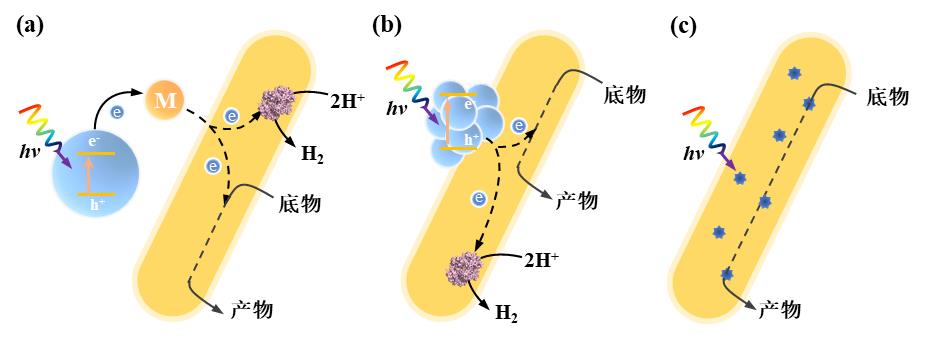
Fig. 2 Material-microbial hybrid forms and corresponding electron transfer mechanisms in light-driven material-microbial hybrid systems. Materials are distributed outside the cell (a), adsorbed on the cell surface (b) or distributed inside the cell (c). (M: electron mediator)
| 产品 种类 | 原料 | 产品 | 材料 | 菌种 | 产品产率 | 量子产率 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 有机酸 | CO2 | 乙酸 | CdS NPs | Moorella thermoacetica | 0.1 mM/h | 2.44 ± 0.62% | [ |
| CdS NPs | Clostridium autoethanogenum | 12.1 mM | - | [ | |||
| AuNCs | Moorella thermoacetica | 6.0 Mm/g/week | 2.86 ± 0.38% | [ | |||
| PFP/PDI | Moorella thermoacetica | 0.6 mM | 1.6% | [ | |||
| MOF | Moorella thermoacetica | - | - | [ | |||
| CdS NPs | Sporomusa ovata | 40.0 mM | 16.8 ± 9% | [ | |||
| CO2 | L-苹果酸 | CdS NPs | Escherichia coli | 1.5 mol/mol | - | [ | |
| 葡萄糖 | 莽草酸 | InP NPs | Saccharomyces cerevisiae | 48.5mg/L | - | [ | |
| 生物醇 | CO2 | 甲醇 | poly(allylamine hydrochloride) | 甲酸脱氢酶、甲醛脱氢酶、酵母醇脱氢酶 | 60.39 μM | - | [ |
| artificial thylakoid | 甲酸脱氢酶、甲醛脱氢酶、酵母醇脱氢酶 | 99 Μm/h | 0.66 ± 0.13% | [ | |||
| CdS-NPs | Clostridium ljungdahlii、 Acetobacterium woodii、 Moorella thermoacetica、 Pseudomonas aeruginosa | 0.4 g/L | - | [ | |||
| CH4 | 甲醇 | BM-TiO2 | Methylosinus trichosporium | 15761.0 μmol/g/h | [ | ||
| 萜类 | TY培养基 | 法尼烯 | meso-Al2O3 | Escherichia coli | 1816.0 mg/L | - | [ |
| CO2/葡萄糖 | 类胡萝卜素 | Au NPs | Chlorella zofingiensis | 10.7 mg/L | - | [ | |
| 聚合物 | CO2/果糖 | PHB | g-C3N4 | Ralstonia eutropha | 9.1 g/L | 29.72% ± 5.53% | [ |
| CO2/果糖 | PHB | CdS NPs | Cupriavidus necator | 0.6 mg/L | - | [ |
Tab. 1 Chemicals synthesized by light-driven material-microbial hybrid system
| 产品 种类 | 原料 | 产品 | 材料 | 菌种 | 产品产率 | 量子产率 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 有机酸 | CO2 | 乙酸 | CdS NPs | Moorella thermoacetica | 0.1 mM/h | 2.44 ± 0.62% | [ |
| CdS NPs | Clostridium autoethanogenum | 12.1 mM | - | [ | |||
| AuNCs | Moorella thermoacetica | 6.0 Mm/g/week | 2.86 ± 0.38% | [ | |||
| PFP/PDI | Moorella thermoacetica | 0.6 mM | 1.6% | [ | |||
| MOF | Moorella thermoacetica | - | - | [ | |||
| CdS NPs | Sporomusa ovata | 40.0 mM | 16.8 ± 9% | [ | |||
| CO2 | L-苹果酸 | CdS NPs | Escherichia coli | 1.5 mol/mol | - | [ | |
| 葡萄糖 | 莽草酸 | InP NPs | Saccharomyces cerevisiae | 48.5mg/L | - | [ | |
| 生物醇 | CO2 | 甲醇 | poly(allylamine hydrochloride) | 甲酸脱氢酶、甲醛脱氢酶、酵母醇脱氢酶 | 60.39 μM | - | [ |
| artificial thylakoid | 甲酸脱氢酶、甲醛脱氢酶、酵母醇脱氢酶 | 99 Μm/h | 0.66 ± 0.13% | [ | |||
| CdS-NPs | Clostridium ljungdahlii、 Acetobacterium woodii、 Moorella thermoacetica、 Pseudomonas aeruginosa | 0.4 g/L | - | [ | |||
| CH4 | 甲醇 | BM-TiO2 | Methylosinus trichosporium | 15761.0 μmol/g/h | [ | ||
| 萜类 | TY培养基 | 法尼烯 | meso-Al2O3 | Escherichia coli | 1816.0 mg/L | - | [ |
| CO2/葡萄糖 | 类胡萝卜素 | Au NPs | Chlorella zofingiensis | 10.7 mg/L | - | [ | |
| 聚合物 | CO2/果糖 | PHB | g-C3N4 | Ralstonia eutropha | 9.1 g/L | 29.72% ± 5.53% | [ |
| CO2/果糖 | PHB | CdS NPs | Cupriavidus necator | 0.6 mg/L | - | [ |
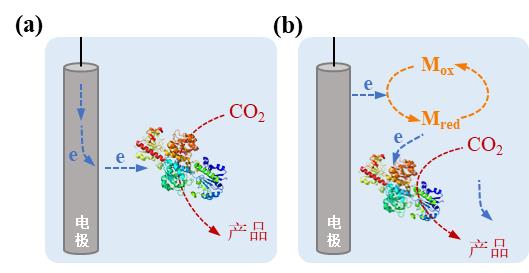
Fig. 3 Enzyme-electrode hybrid forms and corresponding electron transport mechanisms in electrically driven material-microbial hybrid system. (a) Direct electron transfer between cell and electrode; (b) Indirect electron transfer between cell and electrode. (Mox: oxidized mediator, Mred: reduced mediator)
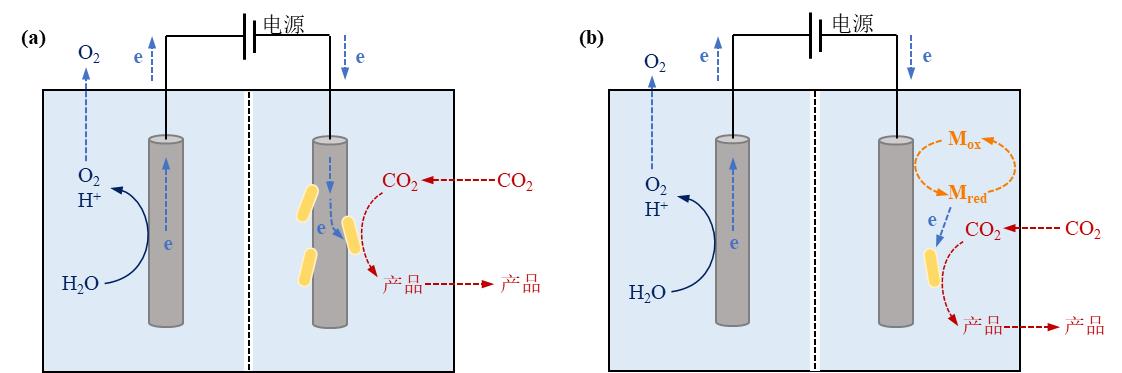
Fig. 4 Microbe-electrode hybrid forms and corresponding electron transport mechanisms in electrically driven material-microbial hybrid system. (a) Direct electron transfer between cell and electrode; (b) Indirect electron transfer between cell and electrode. (Mox: oxidized mediator, Mred: reduced mediator)
| 1 | Wen D, Fang W, Liu Y, et al. Valorization of carbon dioxide with alcohols[J]. Chinese Chemical Letters, 2024, 35(7): 109394. |
| 2 | Su H, Lin J. Biosynthesis pathways of expanding carbon chains for producing advanced biofuels[J]. Biotechnology for Biofuels and Bioproducts, 2023, 16(1): 109. |
| 3 | Zhang C, Ottenheim C, Weingarten M, et al. Microbial utilization of next-generation feedstocks for the biomanufacturing of value-added chemicals and food ingredients[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10. |
| 4 | Bo C, Liu J, Zhang Y, et al. Effective photocatalytic methane oxidation over the TiO2/methanotrophs system[J]. Nano Today, 2023, 52: 101938. |
| 5 | 伊晓峰, 吴见平, 王远鹏. 半导体纳米材料介导微生物固定CO2研究进展[J]. 应用与环境生物学报, 2023, 29(4): 795-802. |
| 6 | Li A, Cao X, Fu R, et al. Biocatalysis of CO2 and CH4: Key enzymes and challenges[J]. Biotechnology Advances, 2024, 72: 108347. |
| 7 | Fang Z, Tang Y J, Koffas M A. Harnessing electrical-to-biochemical conversion for microbial synthesis[J]. Current Opinion in Biotechnology, 2022, 75: 102687. |
| 8 | Li J, Tian Y, Zhou Y, et al. Abiotic-biological hybrid systems for CO2 conversion to value-added chemicals and fuels[J]. Transactions of Tianjin University, 2020, 26(4): 237-247. |
| 9 | Hamby H, Li B, Shinopoulos K E, et al. Light-driven carbon-carbon bond formation via CO2 reduction catalyzed by complexes of CdS nanorods and a 2-oxoacid oxidoreductase[J]. Proceedings of the National Academy of Sciences, 2020, 117(1): 135-140.. |
| 10 | Li J, Han H, Chang Y, et al. The material-microorganism interface in microbial hybrid electrocatalysis systems[J]. Nanoscale, 2023, 15(13): 6009-6024. |
| 11 | Proppe A H, Li Y C, Aspuru-Guzik A, et al. Bioinspiration in light harvesting and catalysis[J]. Nature Reviews Materials, 2020, 5(11): 828-846. |
| 12 | Zavafer A, Cheah M H, Hillier W, et al. Photodamage to the oxygen evolving complex of photosystem II by visible light[J]. Scientific Reports, 2015, 5(1): 16363. |
| 13 | Murata N, Nishiyama Y. ATP is a driving force in the repair of photosystem II during photoinhibition[J]. Plant, Cell & Environment, 2018, 41(2): 285-299. |
| 14 | Liu H, Cheng M, Liu Y, et al. Single atoms meet metal-organic frameworks: collaborative efforts for efficient photocatalysis[J]. Energy & Environmental Science, 2022, 15(9): 3722-3749. |
| 15 | Sun X, Huang H, Zhao Q, et al. Thin-layered photocatalysts[J]. Advanced Functional Materials, 2020, 30(22): 1910005. |
| 16 | Hawkins A S, Mcternan P M, Lian H, et al. Biological conversion of carbon dioxide and hydrogen into liquid fuels and industrial chemicals[J]. Current Opinion in Biotechnology, 2013, 24(3): 376-384. |
| 17 | Yang N, Tian Y, Zhang M, et al. Photocatalyst-enzyme hybrid systems for light-driven biotransformation[J]. Biotechnology Advances, 2022, 54: 107808. |
| 18 | Chen K, Arnold F H. Engineering new catalytic activities in enzymes[J]. Nature Catalysis, 2020, 3(3): 203-213. |
| 19 | Noji T, Jin T, Nango M, et al. CO2 photoreduction by formate dehydrogenase and a Ru-complex in a nanoporous glass reactor[J]. ACS Applied Materials & Interfaces, 2017, 9(4): 3260-3265. |
| 20 | Chaudhary Y S, Woolerton T W, Allen C S, et al. Visible light-driven CO2 reduction by enzyme coupled CdS nanocrystals[J]. Chemical Communications, 2011, 48(1): 58-60. |
| 21 | Woolerton T W, Sheard S, Reisner E, et al. Efficient and clean photoreduction of CO2 to CO by enzyme-modified TiO2 nanoparticles using visible light[J]. Journal of the American Chemical Society, 2010, 132(7): 2132-2133. |
| 22 | Li S, Shi J, Liu S, et al. Molecule-electron-proton transfer in enzyme-photo-coupled catalytic system[J]. Chinese Journal of Catalysis, 2023, 44: 96-110. |
| 23 | Kornienko N, Zhang J Z, Sakimoto K K, et al. Interfacing nature's catalytic machinery with synthetic materials for semi-artificial photosynthesis[J]. Nature Nanotechnology, 2018, 13(10): 890-899. |
| 24 | Cestellos-Blanco S, Zhang H, Kim J M, et al. Photosynthetic semiconductor biohybrids for solar-driven biocatalysis[J]. Nature Catalysis, 2020, 3(3): 245-255. |
| 25 | Sakimoto K K, Wong A B, Yang P D. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production[J]. Science, 2016, 351(6268): 74-77. |
| 26 | Song J, Lin H, Zhao G, et al. Photocatalytic material-microorganism hybrid system and its application—a review[J]. Micromachines, 2022, 13(6): 861. |
| 27 | Honda Y, Watanabe M, Hagiwara H, et al. Inorganic/whole-cell biohybrid photocatalyst for highly efficient hydrogen production from water[J]. Applied Catalysis B: Environmental, 2017, 210: 400-406. |
| 28 | Zhang H, Liu H, Tian Z, et al. Bacteria photosensitized by intracellular gold nanoclusters for solar fuel production[J]. Nature Nanotechnology, 2018, 13(10): 900-905. |
| 29 | Jiang Z, Wang B, Yu J C, et al. AglnS2/In2S3 heterostructure sensitization of Escherichia coli for sustainable hydrogen production[J]. Nano Energy, 2018, 46: 234-240. |
| 30 | Wang J, Chen N, Bian G, et al. Solar-driven overproduction of biofuels in microorganisms[J]. Angewandte Chemie International Edition, 2022, 61(32): e202207132. |
| 31 | Ding Y, Bertram J R, Eckert C, et al. Nanorg microbial factories: light-driven renewable biochemical synthesis using quantum dot-bacteria nanobiohybrids[J]. Journal of the American Chemical Society, 2019, 141(26): 10272-10282. |
| 32 | Kumaravel V, Imam M D, Badreldin A, et al. Photocatalytic hydrogen production: Role of sacrificial reagents on the activity of oxide, carbon, and sulfide catalysts[J]. Catalysts, 2019, 9(3): 276. |
| 33 | Gai P, Yu W, Zhao H, et al. Solar-powered organic semiconductor-bacteria biohybrids for CO2 reduction into acetic acid[J]. Angewandte Chemie International Edition, 2020, 59(18): 7224-7229. |
| 34 | Zhou X, Zeng Y, Tang Y Y, et al. Artificial regulation of state transition for augmenting plant photosynthesis using synthetic light-harvesting polymer materials[J]. Science Advances, 2020, 6(35): eabc5237. |
| 35 | Qi R, Zhao H, Zhou X, et al. In situ synthesis of photoactive polymers on a living cell surface via bio-palladium catalysis for modulating biological functions[J]. Angewandte Chemie International Edition, 2021, 60(11): 5759-5765. |
| 36 | Tremblay P L, Xu M, Chen Y, et al. Nonmetallic abiotic-biological hybrid photocatalyst for visible water splitting and carbon dioxide reduction[J]. iScience, 2020, 23(1): 100784. |
| 37 | Jin S, Jeon Y, Jeon M S, et al. Acetogenic bacteria utilize light-driven electrons as an energy source for autotrophic growth[J]. Proceedings of the National Academy of Sciences, 2021, 118(9): e2020552118. |
| 38 | Ji Z, Zhang H, Liu H, et al. Cytoprotective metal-organic frameworks for anaerobic bacteria[J]. Proceedings of the National Academy of Sciences, 2018, 115(42): 10582-10587. |
| 39 | He Y, Wang S, Han X, et al. Photosynthesis of acetate by Sporomusa ovata–CdS biohybrid system[J]. ACS Applied Materials & Interfaces, 2022, 14(20): 23364-23374. |
| 40 | HU G, LI Z, MA D, et al. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals[J]. Nature Catalysis, 2021, 4(5): 395-406. |
| 41 | Guo J L, Suástegui M, Sakimoto K K, et al. Light-driven fine chemical production in yeast biohybrids[J]. Science, 2018, 362(6416): 813-816. |
| 42 | Ji X, Su Z, Wang P, et al. Integration of artificial photosynthesis system for enhanced electronic energy-transfer efficacy: A case study for solar-energy driven bioconversion of carbon dioxide to methanol[J]. Small, 2016, 12(34): 4753-4762. |
| 43 | Zhang S, Shi J, Sun Y, et al. Artificial thylakoid for the coordinated photoenzymatic reduction of carbon dioxide[J]. ACS Catalysis, 2019, 9(5): 3913-3925. |
| 44 | Kumar M, Sahoo P C, Srikanth S, et al. Photosensitization of electro-active microbes for solar assisted carbon dioxide transformation[J]. Bioresource Technology, 2019, 272: 300-307. |
| 45 | Li X, Sun H, Mao X, et al. Enhanced photosynthesis of carotenoids in microalgae driven by light-harvesting gold nanoparticles[J]. ACS Sustainable Chemistry & Engineering, 2020, 8(20): 7600-7608. |
| 46 | Xu M, Tremblay P L, Ding R, et al. Photo-augmented PHB production from CO2 or fructose by Cupriavidus necator and shape-optimized CdS nanorods[J]. Science of The Total Environment, 2021, 753: 142050. |
| 47 | Sakimoto K K, Zhang S J, Yang P. Cysteine-cystine photoregeneration for oxygenic photosynthesis of acetic acid from CO2 by a tandem inorganic-biological hybrid system[J]. Nano Letters, 2016, 16(9): 5883-5887. |
| 48 | Cestellos-Blanco S, Zhang H, Yang P. Solar-driven carbon dioxide fixation using photosynthetic semiconductor bio-hybrids[J]. Faraday Discussions, 2019, 215(0): 54-65. |
| 49 | Zhang H, Wang L, Pérez-Fortes M, et al. Techno-economic optimization of biomass-to-methanol with solid-oxide electrolyzer[J]. Applied Energy, 2020, 258: 114071. |
| 50 | Sheng Y, Guo F, Guo B, et al. Light-driven CO2 reduction with a surface-displayed enzyme cascade–C3N4 Hybrid[J]. ACS Synthetic Biology, 2023, 12(9): 2715-2724. |
| 51 | Wei W, Sun P, Li Z, et al. A surface-display biohybrid approach to light-driven hydrogen production in air[J]. Science Advances, 2018, 4(2): eaap9253. |
| 52 | Rowe S F, Le Gall G, Ainsworth E V, et al. Light-driven H2 evolution and C=C or C=O bond hydrogenation by Shewanella oneidensis: A versatile strategy for photocatalysis by nonphotosynthetic microorganisms[J]. ACS Catalysis, 2017, 7(11): 7558-7566. |
| 53 | Liu K, Wang F Q, Liu K, et al. Light-driven progesterone production by InP-(M. neoaurum) biohybrid system[J]. Bioresources and Bioprocessing, 2022, 9(1): 93. |
| 54 | Zhao Q, Li Y, Shen B, et al. UiO-66-mediated light-driven regeneration of intracellular NADH in Clostridium tyrobutyricum to strengthen butyrate production[J]. ACS Sustainable Chemistry & Engineering, 2023, 11(8): 3405-3415. |
| 55 | Xiao K, Tsang T H, Sun D, et al. Interfacing iodine-doped hydrothermally carbonized carbon with Escherichia coli through an "add-on" mode for enhanced light-driven hydrogen production[J]. Advanced Energy Materials, 2021, 11(21): 2100291. |
| 56 | Wang B, Jiang Z, Yu J C, et al. Enhanced CO2 reduction and valuable C2+ chemical production by a CdS-photosynthetic hybrid system[J]. Nanoscale, 2019, 11(19): 9296-9301. |
| 57 | Ye J, Yu J, Zhang Y, et al. Light-driven carbon dioxide reduction to methane by Methanosarcina barkeri-CdS biohybrid[J]. Applied Catalysis B: Environmental, 2019, 257: 117916. |
| 58 | Zhou Z, Li Y, Li M, et al. Efficient removal for multiple pollutants via Ag2O/BiOBr heterojunction: A promoted photocatalytic process by valid electron transfer pathway[J]. Chinese Chemical Letters, 2020, 31(10): 2698-2704. |
| 59 | Liang J, Xiao K, Wang X, et al. Revisiting solar energy flow in nanomaterial-microorganism hybrid systems[J]. Chemical Reviews, 2024. |
| 60 | Overa S, Ko B H, Zhao Y, et al. Electrochemical approaches for CO2 conversion to chemicals: A journey toward practical applications[J]. Accounts of Chemical Research, 2022, 55(5): 638-648. |
| 61 | Chiranjeevi P, Bulut M, Breugelmans T, et al. Current trends in enzymatic electrosynthesis for CO2 reduction[J]. Current Opinion in Green and Sustainable Chemistry, 2019, 16: 65-70. |
| 62 | Lu Y C, Song W, An D, et al. Designing compartmentalized hydrogel microparticles for cell encapsulation and scalable 3D cell culture[J]. Journal of Materials Chemistry B, 2014, 3(3): 353-360. |
| 63 | Long C J, He Y H, Guan Z. Emerging strategies for asymmetric synthesis: Combining enzyme promiscuity and photo-/electro-redox catalysis[J]. Asian Journal of Organic Chemistry, 2023, 12(2): e202200685. |
| 64 | Cadoux C, Milton R D. Recent enzymatic electrochemistry for reductive reactions[J]. ChemElectroChem, 2020, 7(9): 1974-1986. |
| 65 | Logan B E, Rossi R, Ragab A, et al. Electroactive microorganisms in bioelectrochemical systems[J]. Nature Reviews Microbiology, 2019, 17(5): 307-319. |
| 66 | 焦子悦, 黄小涵, 郭树奇, 等. 微生物固碳的电子供给策略研究进展[J]. 生物工程学报, 2022, 38(7): 2396-2409. |
| 67 | Bian B, Bajracharya S, Xu J, et al. Microbial electrosynthesis from CO2: Challenges, opportunities and perspectives in the context of circular bioeconomy[J]. Bioresource Technology, 2020, 302: 122863. |
| 68 | Combining metal-microbe and microbe-microbe dual direct electron transfer on Fe( 0)-cathode of bio-electrochemical system to enhance anaerobic digestion of cellulose wastewater[J]. Chinese Chemical Letters, 2022, 33(6): 3106-3112. |
| 69 | Omar B, Abou-Shanab R, El-Gammal M, et al. Simultaneous biogas upgrading and biochemicals production using anaerobic bacterial mixed cultures[J]. Water Research, 2018, 142: 86-95. |
| 70 | Lovley D R. Powering microbes with electricity: direct electron transfer from electrodes to microbes[J]. Environmental Microbiology Reports, 2011, 3(1): 27-35. |
| 71 | Koch C, Harnisch F. What is the essence of microbial electroactivity?[J]. Frontiers in Microbiology, 2016, 7. |
| 72 | Bond D R, Lovley D R. Electricity production by Geobacter sulfurreducens attached to electrodes[J]. Applied and Environmental Microbiology, 2003, 69(3): 1548-1555. |
| 73 | Marsili E, Baron D B, Shikhare I D, et al. Shewanella secretes flavins that mediate extracellular electron transfer[J]. Proceedings of the National Academy of Sciences, 2008, 105(10): 3968-3973. |
| 74 | Harrington T D, Mohamed A, Tran V N, et al. Neutral red-mediated microbial electrosynthesis by Escherichia coli, Klebsiella pneumoniae, and Zymomonas mobilis [J]. Bioresource Technology, 2015, 195: 57-65. |
| 75 | Lai B. Burning questions: Exploring the limits of microbial electrochemical technology for industrial biotechnological applications[J]. Microbial Biotechnology, 2024, 17(1). |
| 76 | Claassens N J, Cotton C A R, Kopljar D, et al. Making quantitative sense of electromicrobial production[J]. Nature Catalysis, 2019, 2(5): 437-447. |
| 77 | Zheng T, Xia C. Electrifying biosynthesis for CO2 upcycling[J]. Trends in Chemistry, 2023, 5(1): 7-10. |
| 78 | Zheng T, Zhang M, Wu L, et al. Upcycling CO2 into energy-rich long-chain compounds via electrochemical and metabolic engineering[J]. Nature Catalysis, 2022, 5(5): 388-396. |
| 79 | Tremblay P L, Zhang T. Electrifying microbes for the production of chemicals[J]. Frontiers in Microbiology, 2015, 6(MAR). |
| 80 | Kang N K, Chau T H T, Lee E Y. Engineered methane biocatalysis: strategies to assimilate methane for chemical production[J]. Current Opinion in Biotechnology, 2024, 85: 103031. |
| 81 | 郭树奇, 焦子悦, 费强. 基于化学品生物合成的嗜甲烷菌人工细胞构建及应用进展[J]. 合成生物学, 2021, 2(6): 1017-1029. |
| 82 | Kracke F, Deutzmann J S, Gu W, et al. In situ electrochemical H2 production for efficient and stable power-to-gas electromethanogenesis[J]. Green Chemistry, 2020, 22(18): 6194-6203. |
| 83 | Bai Y, Zhou L, Irfan M, et al. Bioelectrochemical methane production from CO2 by Methanosarcina barkeri via direct and H2-mediated indirect electron transfer[J]. Energy, 2020, 210: 118445. |
| 84 | Fu Q, Kuramochi Y, Fukushima N, et al. Bioelectrochemical analyses of the development of a Thermophilic Biocathode catalyzing electromethanogenesis[J]. Environmental Science & Technology, 2015, 49(2): 1225-1232. |
| 85 | Fixen K R, Zheng Y, Harris D F, et al. Light-driven carbon dioxide reduction to methane by nitrogenase in a photosynthetic bacterium[J]. Proceedings of the National Academy of Sciences, 2016, 113(36): 10163-10167. |
| 86 | Bajracharya S, Srikanth S, Mohanakrishna G, et al. Biotransformation of carbon dioxide in bioelectrochemical systems: State of the art and future prospects[J]. Journal of Power Sources, 2017, 356: 256-273. |
| 87 | Flexer V, Chen J, Donose B C, et al. The nanostructure of three-dimensional scaffolds enhances the current density of microbial bioelectrochemical systems[J]. Energy & Environmental Science, 2013, 6(4): 1291-1298. |
| 88 | Katuri K P, Kamireddy S, Kavanagh P, et al. Electroactive biofilms on surface functionalized anodes: The anode respiring behavior of a novel electroactive bacterium, Desulfuromonas acetexigens [J]. Water Research, 2020, 185: 116284. |
| 89 | Wang X, Li X, Zhu Q. Electrocatalytic nanomaterials improve microbial extracellular electron transfer: A review[J]. Applied Sciences, 2024, 14(15): 6733. |
| 90 | Hou X, Huang L, Zhou P. Synergetic interaction of magnetic field and loaded magnetite for enhanced acetate production in biocathode of microbial electrosynthesis system[J]. International Journal of Hydrogen Energy, 2021, 46(10): 7183-7194. |
| 91 | Zhang T, Nie H, Bain T S, et al. Improved cathode materials for microbial electrosynthesis[J]. Energy & Environmental Science, 2012, 6(1): 217-224. |
| 92 | Tremblay P L, Höglund D, Koza A, et al. Adaptation of the autotrophic acetogen Sporomusa ovata to methanol accelerates the conversion of CO2 to organic products[J]. Scientific Reports, 2015, 5(1): 16168. |
| 93 | Liu C, Colón B C, Ziesack M, et al. Water splitting-biosynthetic system with CO2 reduction efficiencies exceeding photosynthesis[J]. Science, 2016, 352(6290): 1210-1213. |
| 94 | Treece T R, Pattanayak S, Matson M M, et al. Electrical-biological hybrid system for carbon efficient isobutanol production[J]. Metabolic Engineering, 2023, 80: 142-150. |
| 95 | Lim J, Choi S Y, Lee J W, et al. Biohybrid CO2 electrolysis for the direct synthesis of polyesters from CO2 [J]. Proceedings of the National Academy of Sciences, 2023, 120(14): e2221438120. |
| 96 | Liu G, Zhong Y, Liu Z, et al. Solar-driven sugar production directly from CO2 via a customizable electrocatalytic–biocatalytic flow system[J]. Nature Communications, 2024, 15(1): 2636. |
| 97 | Zhang Y, Feng T, Zhou X, et al. Photoelectrocatalytic-microbial biohybrid for nitrogen reduction[J]. Advanced Materials, n/a(n/a): 2407239. |
| 98 | Su Y, Cestellos-Blanco S, Kim J M, et al. Close-packed nanowire-bacteria hybrids for efficient solar-driven CO2 fixation[J]. Joule, 2020, 4(4): 800-811. |
| 99 | Cestellos-Blanco S, Chan R R, Shen Y X, et al. Photosynthetic biohybrid coculture for tandem and tunable CO2 and N2 fixation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(26): e2122364119. |
| 100 | Liu C, Gallagher J J, Sakimoto K K, et al. Nanowire-bacteria hybrids for unassisted solar carbon dioxide fixation to value-added chemicals[J]. Nano letters, 2015, 15(5): 3634. |
| [1] | Shouqi ZHANG, Tao WANG, Yao KONG, Jiasheng ZOU, Yuanning LIU, Zhengren XU. Chemoenzymatic synthesis of natural products: evolution of synthetic methodology and strategy [J]. Synthetic Biology Journal, 2024, 5(5): 913-940. |
| [2] | Yu FU, Fangrui ZHONG. Recent advances in chemically driven enantioselective photobiocatalysis [J]. Synthetic Biology Journal, 2024, 5(5): 1021-1049. |
| [3] | Yanping QI, Jin ZHU, Kai ZHANG, Tong LIU, Yajie WANG. Recent development of directed evolution in protein engineering [J]. Synthetic Biology Journal, 2022, 3(6): 1081-1108. |
| [4] | Huibin WANG, Changli CHE, Song YOU. Recent advances of enzymatic synthesis of organohalogens catalyzed by Fe/αKG-dependent halogenases [J]. Synthetic Biology Journal, 2022, 3(3): 545-566. |
| [5] | Yujiao LOU, Jian XU, Qi WU. Progress of biocatalytic deuteration of inert carbon-hydrogen bonds [J]. Synthetic Biology Journal, 2022, 3(3): 530-544. |
| [6] | Lu YANG, Xudong QU. Application of imine reductase in the synthesis of chiral amines [J]. Synthetic Biology Journal, 2022, 3(3): 516-529. |
| [7] | Liangbin XIONG, Lu SONG, Yunqiu ZHAO, Kun LIU, Yongjun LIU, Fengqing WANG, Dongzhi WEI. Green biomanufacturing of steroids: from biotransformation to de novo synthesis by microorganisms [J]. Synthetic Biology Journal, 2021, 2(6): 942-963. |
| [8] | Faguang ZHANG, Ge QU, Zhoutong SUN, Jun′an MA. From chemical synthesis to biosynthesis: trends toward total synthesis of natural products [J]. Synthetic Biology Journal, 2021, 2(5): 674-696. |
| [9] | Heng TANG, Xin HAN, Shuping ZOU, Yuguo ZHENG. Application of multi-enzyme catalytic system in the synthesis of pharmaceutical chemicals [J]. Synthetic Biology Journal, 2021, 2(4): 559-576. |
| [10] | Shuke WU, Yi ZHOU, Wen WANG, Wei ZHANG, Pengfei GAO, Zhi LI. From single-enzyme catalysis to multienzyme cascade: inspired from Professor Daniel I.C. Wang’s pioneer work in enzyme technology [J]. Synthetic Biology Journal, 2021, 2(4): 543-558. |
| [11] | Junting WANG, Xiaojia GUO, Qing LI, Li WAN, Zongbao ZHAO. Creation of non-natural cofactor-dependent methanol dehydrogenase [J]. Synthetic Biology Journal, 2021, 2(4): 651-661. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||