Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (2): 335-351.DOI: 10.12211/2096-8280.2021-055
• Invited Review • Previous Articles Next Articles
Bioorthogonal functionalization of viruses for biomedical applications
HUANG Lili1, ZHANG Han2, WANG Weiwei1, XIE Haiyan1,2
- 1.Institute of Engineering Medicine,Beijing Institute of Technology,Beijing 100081,China
2.School of Life Science,Beijing Institute of Technology,Beijing 100081,China
-
Received:2021-05-03Revised:2021-08-27Online:2022-05-11Published:2022-04-30 -
Contact:XIE Haiyan
基于生物正交反应的病毒功能化及其生物医学应用
黄利利1, 张韩2, 王伟伟1, 谢海燕1,2
- 1.北京理工大学医工融合研究院,北京 100081
2.北京理工大学生命学院,北京 100081
-
通讯作者:谢海燕 -
作者简介:黄利利 (1984—),女,博士,特别研究员,博士生导师。研究方向:建立病毒的荧光标记新方法,并实时动态示踪其侵染过程,为诠释病毒的感染机制和相应药物的研发提供依据;构建具有肿瘤诊疗一体化功能的溶瘤病毒纳米颗粒,推动溶瘤病毒的临床应用。 E-mail:llhuang@bit.edu.cn谢海燕 (1975—),女,博士,教授,博士生导师。研究方向:仿生生物探针构建及其活体成像应用;智能纳米生物医学诊疗系统构建与应用;溶瘤病毒工程化改造。 E-mail:hyanxie@bit.edu.cn -
基金资助:国家自然科学基金(21904012);国家杰出青年科学基金(22025401)
CLC Number:
Cite this article
HUANG Lili, ZHANG Han, WANG Weiwei, XIE Haiyan. Bioorthogonal functionalization of viruses for biomedical applications[J]. Synthetic Biology Journal, 2022, 3(2): 335-351.
黄利利, 张韩, 王伟伟, 谢海燕. 基于生物正交反应的病毒功能化及其生物医学应用[J]. 合成生物学, 2022, 3(2): 335-351.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-055

Fig. 1 Bioorthogonal reactions based on aldehydes or ketones[13][Aldehydes or ketones can condense with aminooxy compounds (top) or hydrazide compounds (bottom) to form stable oxime or hydrazine linkages, respectively]
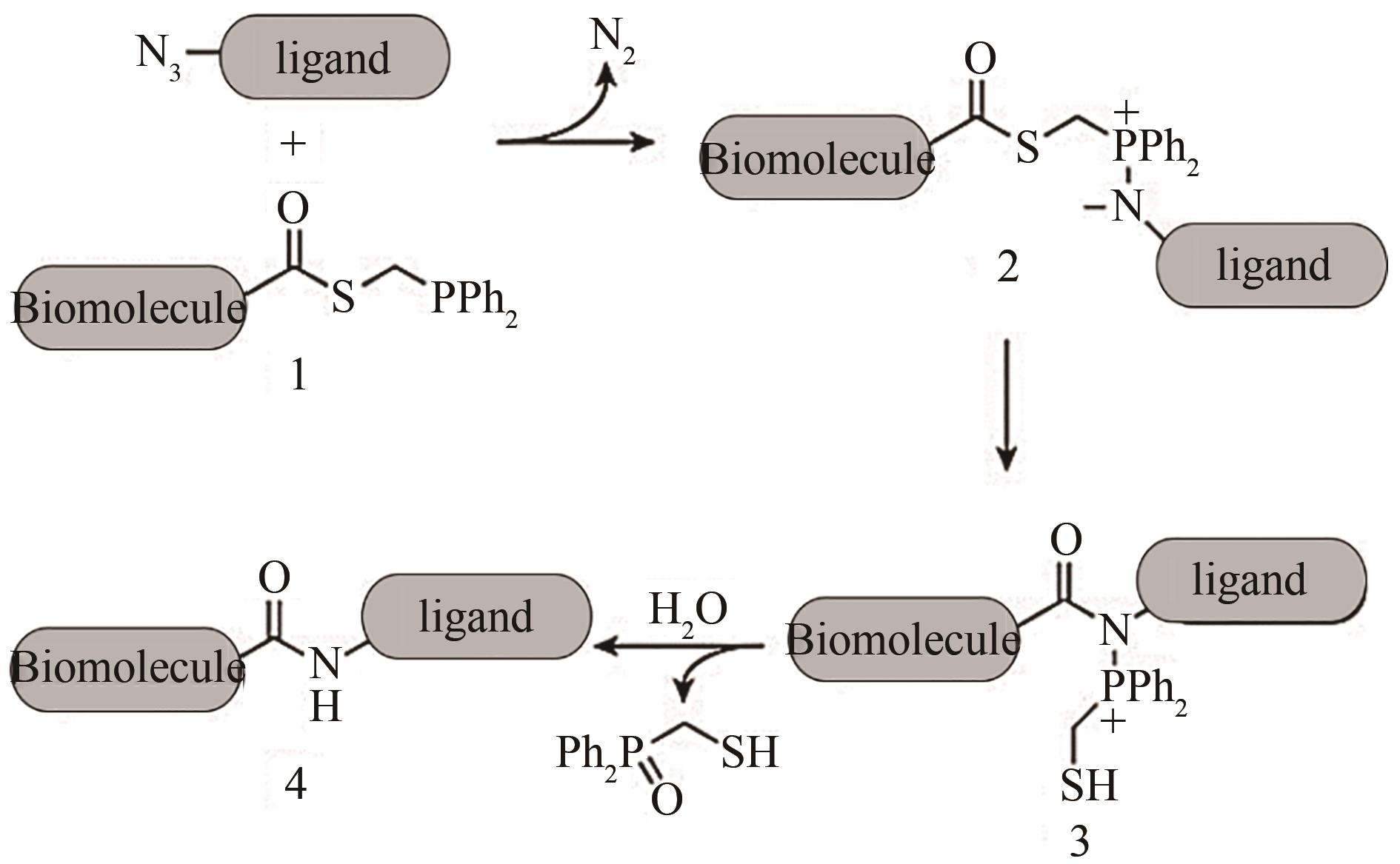
Fig. 2 Biomacromolecules coupling with ligands by the traceless Staudinger ligation reaction[13].[The phosphine-modified biomacromolecule (1) could react with the azide-linked ligand by the Staudinger ligation reaction to yield iminophosphorane (2), which rearranges to produce the intermediate (3) for hydrolysis to generate the coupling product (4) and a phosphine oxide by-product.]
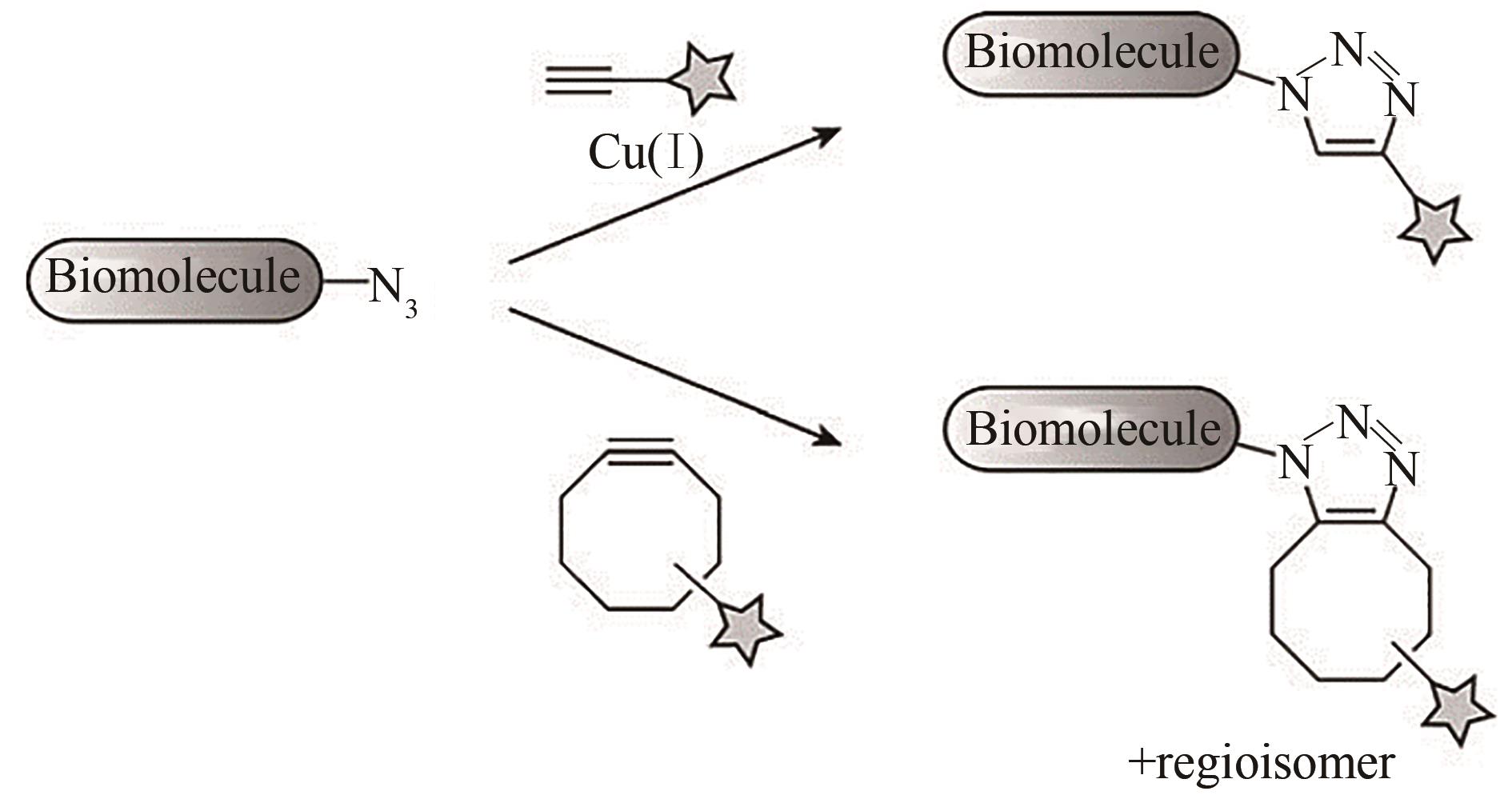
Fig. 3 Bioorthogonal cycloadditions of azides and alkynes to form triazoles[13][Terminal alkynes are activated by Cu (Ⅰ) to undergo cycloaddition with azides (top). Cyclooctynes react with azides through a strain-promoted cycloaddition (bottom).]
Fig. 4 Site-specific in vivo labeling of enveloped influenza VLPs[71]During intracellular protein synthesis, the AHA is incorporated into the nascent HA proteins. The modified HA is further incorporated into the vesicles’ envelope that can secret from the cells, carrying the chemical modification. Addition of the Alexa 488-cyclooctyne reagent allows the site-specific modification of HA by strain-promoted alkyne-azide cycloaddition(SPAAC)
Fig. 5 Modification of an enveloped measles virus can be achieved by the metabolic incorporation of azido sugars into the host cells through the protein glycosylation process[85]
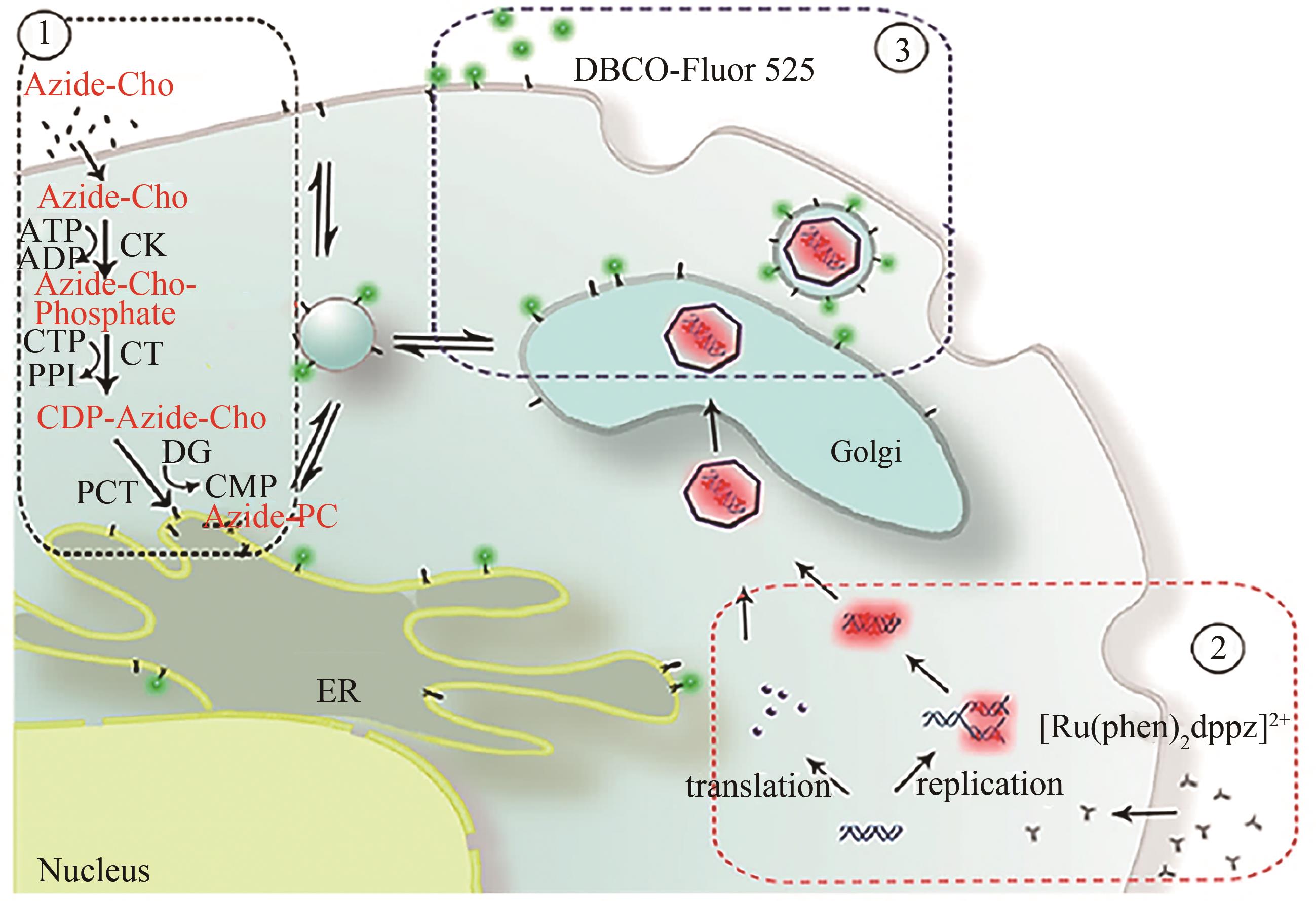
Fig. 6 Dual-fluorescent labeling of the viral envelope and nucleic acid in host cells[91] [The vaccinia virus (VACV) propagates in the presence of both azide-Cho and [Ru(phen)2(dppz)]2+ in the host cell. The biosynthesis and incorporation of azide-Cho-containing phospholipids in the host cells are carried out at first (①);then the cells are infected with VACV, and the [Ru(phen)2(dppz)]2+ is added into the medium at 2 h after the infection, which could enter the cells through the permeable cytomembrane due to the cytopathogenic effect of the virus to label the nucleic acid of the virions (②); at 24 h after the infection, DBCO-Fluor 525 is added into the medium to label the envelope of the VACV (③); after another 24 h with the infection, the dual-labeled virions are finally assembled and released.]
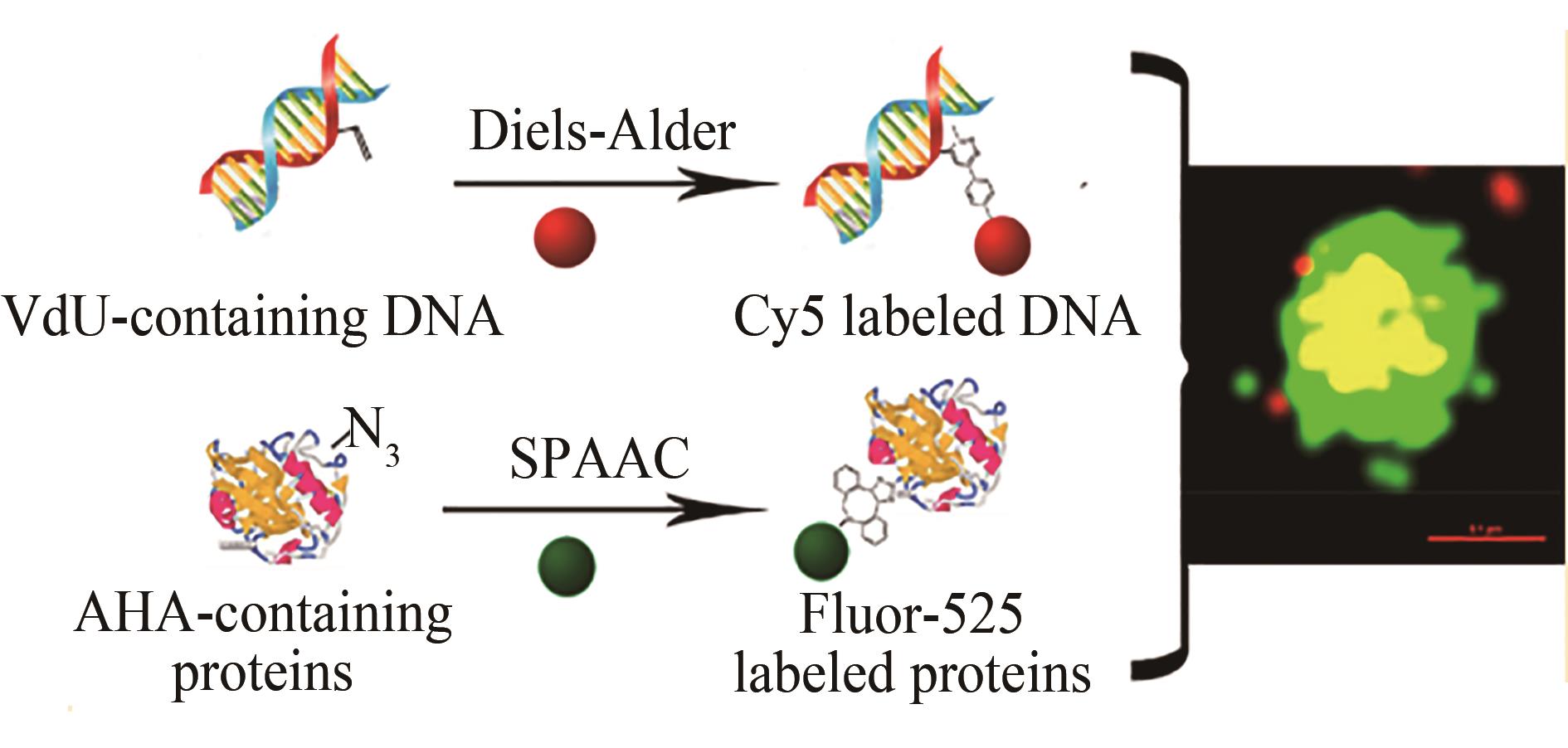
Fig. 7 Dual-fluorescent labeling of viral protein and nucleic acid in host cells[99](AHA-containing protein is labeled by SPAAC reaction,and the VdU-containing nucleic acid is labeled by IEDDA reaction)
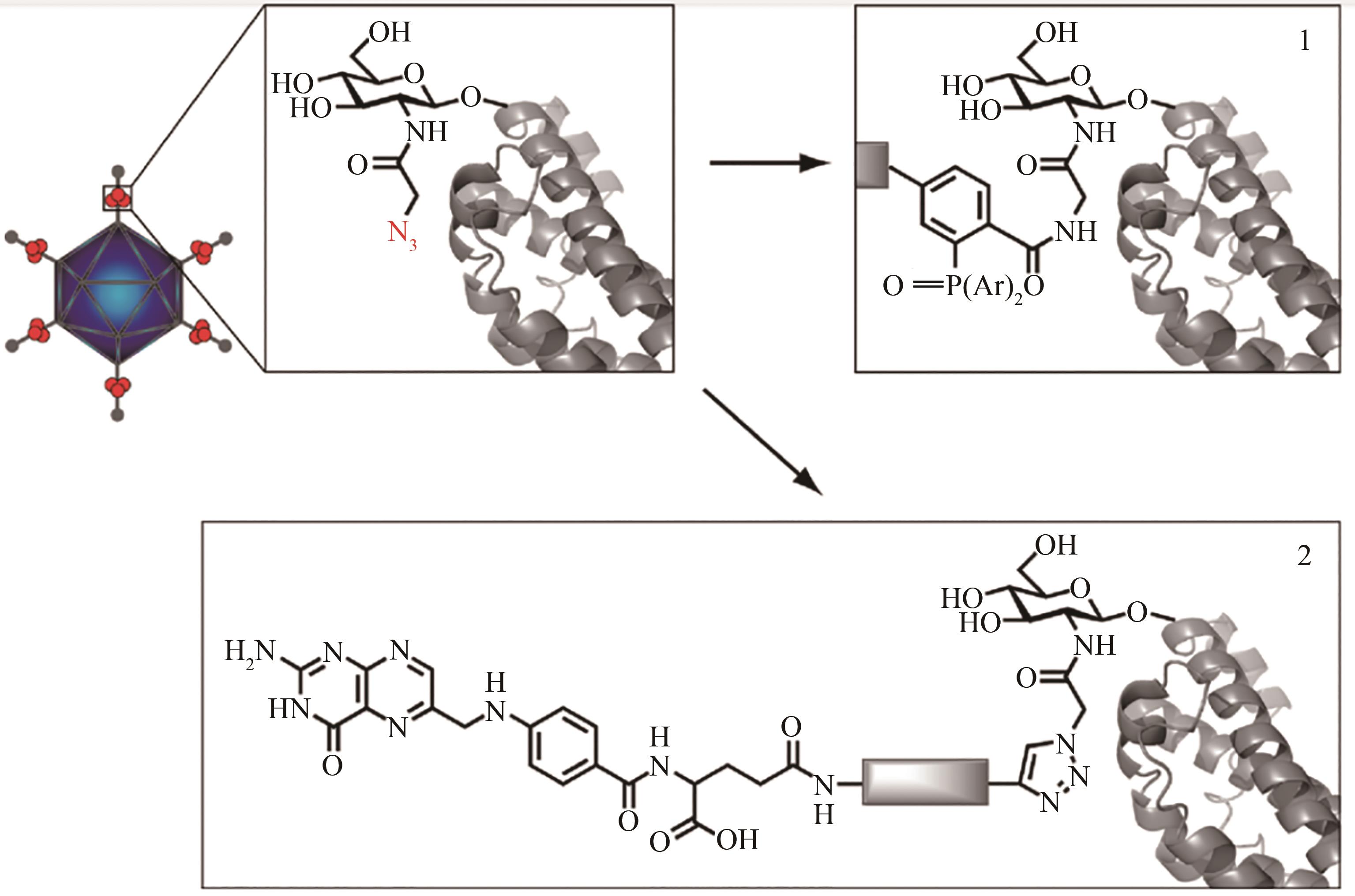
Fig. 8 A cartoon illustration for incorporating the O-GlcNAz residue with the adenoviral fiber protein and subsequent chemical modification with the ligand[84][Potential sites for the O-GlcNAz incorporation are indicated with red circles. Either “Click”(1)or Staudinger ligation(2)chemistry is used to decorate metabolically labeled adenovirus.]
| 1 | EDWARDSON T G W, HILVERT D. Virus-inspired function in engineered protein cages[J]. Journal of the American Chemical Society, 2019, 141(24): 9432-9443. |
| 2 | MATSUURA K. Synthetic approaches to construct viral capsid-like spherical nanomaterials[J]. Chemical Communications, 2018, 54(65): 8944-8959. |
| 3 | SHERMAN M B, GUENTHER R H, TAMA F, et al. Removal of divalent cations induces structural transitions in red clover necrotic mosaic virus, revealing a potential mechanism for RNA release[J]. Journal of Virology, 2006, 80(21): 10395-10406. |
| 4 | KENDALL A, MCDONALD M, BIAN W, et al. Structure of flexible filamentous plant viruses[J]. Journal of Virology, 2008, 82(19): 9546-9554. |
| 5 | SACHSE C, CHEN J Z, COUREUX P D, et al. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus[J]. Journal of Molecular Biology, 2007, 371(3): 812-835. |
| 6 | PEEK L J, MIDDAUGH C R, BERKLAND C. Nanotechnology in vaccine delivery[J]. Advanced Drug Delivery Reviews, 2008, 60(8): 915-928. |
| 7 | PEJAWAR-GADDY S, RAJAWAT Y, HILIOTI Z, et al. Generation of a tumor vaccine candidate based on conjugation of a MUC1 peptide to polyionic papillomavirus virus-like particles[J]. Cancer Immunology, Immunotherapy: CII, 2010, 59(11): 1685-1696. |
| 8 | EDELSTEIN M L, ABEDI M R, WIXON J, et al. Gene therapy clinical trials worldwide 1989—2004—an overview[J]. The Journal of Gene Medicine, 2004, 6(6): 597-602. |
| 9 | SAXON E, BERTOZZI C R. Cell surface engineering by a modified Staudinger reaction[J]. Science, 2000, 287(5460): 2007-2010. |
| 10 | 汪欣, 张贤睿, 黄宗煜, 等. 生物正交反应在我国的研究进展[J]. 化学学报, 2021, 79(4): 406-413. |
| WANG X, ZHANG X R, HUANG Z Y, et al. Recent progress of bioorthogonal chemistry in China[J]. Acta Chimica Sinica, 2021, 79(4): 406-413. | |
| 11 | CAÑEQUE T, MÜLLER S, RODRIGUEZ R. Visualizing biologically active small molecules in cells using click chemistry[J]. Nature Reviews Chemistry, 2018, 2(9): 202-215. |
| 12 | GAMA M A S, FILHO H G B, BIZZO H R, et al. Analytical shortcomings and other considerations related to the identification of biomarkers of dairy fat intake[J]. European Journal of Clinical Nutrition, 2017, 71(8): 1022-1023. |
| 13 | SLETTEN E, BERTOZZI C. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality[J]. Angewandte Chemie International Edition, 2009, 48(38): 6974-6998. |
| 14 | JENCKS W P. Studies on the mechanism of oxime and semicarbazone formation1[J]. Journal of the American Chemical Society, 1959, 81(2): 475-481. |
| 15 | CARRICO I S, CARLSON B L, BERTOZZI C R. Introducing genetically encoded aldehydes into proteins[J]. Nature Chemical Biology, 2007, 3(6): 321-322. |
| 16 | CORNISH V W, HAHN K M, SCHULTZ P G. Site-specific protein modification using a ketone handle[J]. Journal of the American Chemical Society, 1996, 118(34): 8150-8151. |
| 17 | ROSE K, VIZZAVONA J. Stepwise solid-phase synthesis of polyamides as linkers[J]. Journal of the American Chemical Society, 1999, 121(30): 7034-7038. |
| 18 | O'SHANNESSY D J, DOBERSEN M J, QUARLES R H. A novel procedure for labeling immunoglobulins by conjugation to oligosaccharide moieties[J]. Immunology Letters, 1984, 8(5): 273-277. |
| 19 | ZHANG Z W, SMITH B A C, WANG L, et al. A new strategy for the site-specific modification of proteins in vivo [J]. Biochemistry, 2003, 42(22): 6735-6746. |
| 20 | GRIFFIN R J. 3 the medicinal chemistry of the azido group[J]. Progress in Medicinal Chemistry, 1994, 31: 121-232. |
| 21 | HENDRICKS S B, PAULING L. The crystal structures of sodium and potassium trinitrides and potassium cyanate and the nature of the trinitride group[J]. Journal of the American Chemical Society, 1925, 47(12): 2904-2920. |
| 22 | SIDGWICK N V, SUTTON L E, THOMAS W. 107. Dipole moments and structures of the organic azides and aliphatic diazo-compounds[J]. Journal of the Chemical Society, 1933: 406. |
| 23 | KNAGGS I E. Crystal structure of cyanuric triazide[J]. Nature, 1935, 135(3407): 268. |
| 24 | HANGAUER M, BERTOZZI C. A FRET-based fluorogenic phosphine for live-cell imaging with the Staudinger ligation [J]. Angewandte Chemie International Edition, 2008, 47(13): 2394-2397. |
| 25 | TAM A, SOELLNER M B, RAINES R T. Water-soluble phosphinothiols for traceless Staudinger ligation and integration with expressed protein ligation [J]. Journal of the American Chemical Society, 2007, 129(37): 11421-11430. |
| 26 | MOSES J E, MOORHOUSE A D. The growing applications of click chemistry [J]. Chemical Society Reviews, 2007, 36(8): 1249-1262. |
| 27 | ROSTOVTSEV V V, GREEN L G, FOKIN V V, et al. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective "ligation" of azides and terminal alkynes[J]. Angewandte Chemie International Edition, 2002, 41(14): 2596-2599. |
| 28 | TORNØE C W, CHRISTENSEN C, MELDAL M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides[J]. The Journal of Organic Chemistry, 2002, 67(9): 3057-3064. |
| 29 | RODIONOV V O, PRESOLSKI S I, DÍAZ D D, et al. Ligand-accelerated Cu-catalyzed azide-alkyne cycloaddition: a mechanistic report[J]. Journal of the American Chemical Society, 2007, 129(42): 12705-12712. |
| 30 | CHAN T R, HILGRAF R, SHARPLESS K B, et al. Polytriazoles as copper(I)-stabilizing ligands in catalysis[J]. Organic Letters, 2004, 6(17): 2853-2855. |
| 31 | MACKENZIE D A, PEZACKI J P. Kinetics studies of rapid strain-promoted [3+2] cycloadditions of nitrones with bicyclo[6.1.0]nonyne[J]. Canadian Journal of Chemistry, 2014, 92(4): 337-340. |
| 32 | JACOBS C L, GOON S, YAREMA K J, et al. Substrate specificity of the sialic acid biosynthetic pathway[J]. Biochemistry, 2001, 40(43): 12864-12874. |
| 33 | HONG V, PRESOLSKI S, MA C L, et al. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation[J]. Angewandte Chemie International Edition, 2009, 48(52): 9879-9883. |
| 34 | BESANCENEY-WEBLER C, JIANG H, ZHENG T Q, et al. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: a comparative study[J]. Angewandte Chemie International Edition, 2011, 50(35): 8051-8056. |
| 35 | SORIANO DEL AMO D, WANG W, JIANG H, et al. Biocompatible copper(I) catalysts for in vivo imaging of glycans[J]. Journal of the American Chemical Society, 2010, 132(47): 16893-16899. |
| 36 | AGARD N J, PRESCHER J A, BERTOZZI C R. A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems[J]. Journal of the American Chemical Society, 2004, 126(46): 15046-15047. |
| 37 | CODELLI J A, BASKIN J M, AGARD N J, et al. Second-generation difluorinated cyclooctynes for copper-free click chemistry[J]. Journal of the American Chemical Society, 2008, 130(34): 11486-11493. |
| 38 | SLETTEN E M, BERTOZZI C R. A hydrophilic azacyclooctyne for Cu-free click chemistry[J]. Organic Letters, 2008, 10(14): 3097-3099. |
| 39 | NING X H, GUO J, WOLFERT M, et al. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions[J]. Angewandte Chemie International Edition, 2008, 47(12): 2253-2255. |
| 40 | MCKAY C S, FINN M G. Click chemistry in complex mixtures: bioorthogonal bioconjugation[J]. Chemistry & Biology, 2014, 21(9): 1075-1101. |
| 41 | BASKIN J M, PRESCHER J A, LAUGHLIN S T, et al. Copper-free click chemistry for dynamic in vivo imaging[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(43): 16793-16797. |
| 42 | LAUGHLIN S T, BASKIN J M, AMACHER S L, et al. In vivo imaging of membrane-associated glycans in developing zebrafish[J]. Science, 2008, 320(5876): 664-667. |
| 43 | DEHNERT K W, BASKIN J M, LAUGHLIN S T, et al. Imaging the sialome during zebrafish development with copper-free click chemistry[J]. ChemBioChem, 2012, 13(3): 353-357. |
| 44 | LAUGHLIN S T, BERTOZZI C R. In vivo imaging of caenorhabditis elegans glycans[J]. ACS Chemical Biology, 2009, 4(12): 1068-1072. |
| 45 | CHANG P V, PRESCHER J A, SLETTEN E M, et al. Copper-free click chemistry in living animals[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(5): 1821-1826. |
| 46 | WU H X, DEVARAJ N K. Inverse electron-demand Diels-Alder bioorthogonal reactions[J]. Topics in Current Chemistry, 2016, 374(1): 3. |
| 47 | BLACKMAN M L, ROYZEN M, FOX J M. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity[J]. Journal of the American Chemical Society, 2008, 130(41): 13518-13519. |
| 48 | DEVARAJ N K, WEISSLEDER R, HILDERBRAND S A. Tetrazine-based cycloadditions: application to pretargeted live cell imaging[J]. Bioconjugate Chemistry, 2008, 19(12): 2297-2299. |
| 49 | LANG K, DAVIS L, TORRES-KOLBUS J, et al. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction[J]. Nature Chemistry, 2012, 4(4): 298-304. |
| 50 | DEVARAJ N K, WEISSLEDER R. Biomedical applications of tetrazine cycloadditions[J]. Accounts of Chemical Research, 2011, 44(9): 816-827. |
| 51 | XIONG D C, ZHU J J, HAN M J, et al. Rapid probing of sialylated glycoproteins in vitro and in vivo via metabolic oligosaccharide engineering of a minimal cyclopropene reporter[J]. Organic & Biomolecular Chemistry, 2015, 13(13): 3911-3917. |
| 52 | PATTERSON D M, NAZAROVA L A, XIE B, et al. Functionalized cyclopropenes as bioorthogonal chemical reporters[J]. Journal of the American Chemical Society, 2012, 134(45): 18638-18643. |
| 53 | RIEDER U, LUEDTKE N W. Alkene-tetrazine ligation for imaging cellular DNA[J]. Angewandte Chemie International Edition, 2014, 53(35): 9168-9172. |
| 54 | BEST M D. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules[J]. Biochemistry, 2009, 48(28): 6571-6584. |
| 55 | HAUN J B, DEVARAJ N K, MARINELLI B S, et al. Probing intracellular biomarkers and mediators of cell activation using nanosensors and bioorthogonal chemistry[J]. ACS Nano, 2011, 5(4): 3204-3213. |
| 56 | STEINMETZ N F, HONG V, SPOERKE E D, et al. Buckyballs meet viral nanoparticles: candidates for biomedicine[J]. Journal of the American Chemical Society, 2009, 131(47): 17093-17095. |
| 57 | PRASUHN D E JR, YEH R M, OBENAUS A, et al. Viral MRI contrast agents: coordination of Gd by native virions and attachment of Gd complexes by azide-alkyne cycloaddition[J]. Chemical Communications, 2007(12): 1269-1271. |
| 58 | STEINMETZ N F, MERTENS M E, TAUROG R E, et al. Potato virus X as a novel platform for potential biomedical applications[J]. Nano Letters, 2010, 10(1): 305-312. |
| 59 | HAO J, HUANG L L, ZHANG R, et al. A mild and reliable method to label enveloped virus with quantum dots by copper-free click chemistry[J]. Analytical Chemistry, 2012, 84(19): 8364-8370. |
| 60 | HOOKER J M, DATTA A, BOTTA M, et al. Magnetic resonance contrast agents from viral capsid shells: a comparison of exterior and interior cargo strategies[J]. Nano Letters, 2007, 7(8): 2207-2210. |
| 61 | DIRKSEN A, DAWSON P E. Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling[J]. Bioconjugate Chemistry, 2008, 19(12): 2543-2548. |
| 62 | BRUNEL F M, LEWIS J D, DESTITO G, et al. Hydrazone ligation strategy to assemble multifunctional viral nanoparticles for cell imaging and tumor targeting[J]. Nano Letters, 2010, 10(3): 1093-1097. |
| 63 | GILMORE J, SCHECK R, ESSER-KAHN A, et al. N-terminal protein modification through a biomimetic transamination reaction[J]. Angewandte Chemie International Edition, 2008, 47(41): 7788. |
| 64 | CARRICO Z M, FARKAS M E, ZHOU Y, et al. N-Terminal labeling of filamentous phage to create cancer marker imaging agents[J]. ACS Nano, 2012, 6(8): 6675-6680. |
| 65 | KOVACS E W, HOOKER J M, ROMANINI D W, et al. Dual-surface-modified bacteriophage MS2 as an ideal scaffold for a viral capsid-based drug delivery system[J]. Bioconjugate Chemistry, 2007, 18(4): 1140-1147. |
| 66 | LAKADAMYALI M, RUST M J, BABCOCK H P, et al. Visualizing infection of individual influenza viruses[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(16): 9280-9285. |
| 67 | 王芳芳, 潘宏, 何华美, 等. 基于代谢工程与生物正交反应的病毒原位标记技术研究[J]. 集成技术, 2019, 8(3): 1-9. |
| WANG F F, PAN H, HE H M, et al. Research on virus in situ labeling technology based on metabolic engineering and bioorthogonal reaction[J]. Journal of Integration Technology, 2019, 8(3): 1-9. | |
| 68 | XIE R, HONG S L, CHEN X. Cell-selective metabolic labeling of biomolecules with bioorthogonal functionalities[J]. Current Opinion in Chemical Biology, 2013, 17(5): 747-752. |
| 69 | WANG F F, PAN H, YAO X J, et al. Bioorthogonal metabolic labeling utilizing protein biosynthesis for dynamic visualization of nonenveloped enterovirus 71 infection[J]. ACS Applied Materials & Interfaces, 2020, 12(3): 3363-3370. |
| 70 | KRALL N, CRUZ F P DA, BOUTUREIRA O, et al. Site-selective protein-modification chemistry for basic biology and drug development[J]. Nature Chemistry, 2016, 8(2): 103-113. |
| 71 | CARVALHO S B, FREIRE J M, MOLEIRINHO M G, et al. Bioorthogonal strategy for bioprocessing of specific-site-functionalized enveloped influenza-virus-like particles[J]. Bioconjugate Chemistry, 2016, 27(10): 2386-2399. |
| 72 | STRABLE E, PRASUHN D E JR, UDIT A K, et al. Unnatural amino acid incorporation into virus-like particles[J]. Bioconjugate Chemistry, 2008, 19(4): 866-875. |
| 73 | 李劼, 王杰, 陈鹏. 基于非天然氨基酸的蛋白质生物正交标记[J]. 化学学报,2012, 70: 1439-1445. |
| LI J, WANG J, CHEN P. Unnatural amino acid mediated protein bioorthogonal labeling [J]. Acta Chimica Sinica, 2012, 70: 1439-1445. | |
| 74 | WANG L, BROCK A, HERBERICH B, et al. Expanding the genetic code of Escherichia coli [J]. Science, 2001, 292(5516): 498-500. |
| 75 | CARRICO Z M, ROMANINI D W, MEHL R A, et al. Oxidative coupling of peptides to a virus capsid containing unnatural amino acids[J]. Chemical Communications, 2008(10): 1205-1207. |
| 76 | TONG G J, HSIAO S C, CARRICO Z M, et al. Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles[J]. Journal of the American Chemical Society, 2009, 131(31): 11174-11178. |
| 77 | STEPHANOPOULOS N, CARRICO Z, FRANCIS M. Nanoscale integration of sensitizing chromophores and porphyrins with bacteriophage MS2[J]. Angewandte Chemie International Edition, 2009, 48(50): 9498-9502. |
| 78 | LIN S X, YAN H, LI L, et al. Site-specific engineering of chemical functionalities on the surface of live Hepatitis D virus[J]. Angewandte Chemie International Edition, 2013, 52(52): 13970-13974. |
| 79 | SAKIN V, HANNE J, DUNDER J, et al. A versatile tool for live-cell imaging and super-resolution nanoscopy studies of HIV-1 env distribution and mobility[J]. Cell Chemical Biology, 2017, 24(5): 635-645.e5. |
| 80 | ROUHANIFARD S H, NORDSTRØM L U, ZHENG T Q, et al. Chemical probing of glycans in cells and organisms[J]. Chemical Society Reviews, 2013, 42(10): 4284-4296. |
| 81 | MAHAL L K, YAREMA K J, BERTOZZI C R. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis[J]. Science, 1997, 276(5315): 1125-1128. |
| 82 | YOON H Y, KOO H, KIM K, et al. Molecular imaging based on metabolic glycoengineering and bioorthogonal click chemistry[J]. Biomaterials, 2017, 132: 28-36. |
| 83 | SLETTEN E M, BERTOZZI C R. From mechanism to mouse: a tale of two bioorthogonal reactions[J]. Accounts of Chemical Research, 2011, 44(9): 666-676. |
| 84 | BANERJEE P S, OSTAPCHUK P, HEARING P, et al. Chemoselective attachment of small molecule effector functionality to human adenoviruses facilitates gene delivery to cancer cells[J]. Journal of the American Chemical Society, 2010, 132(39): 13615-13617. |
| 85 | ZHAO X, CAI L, ADOGLA E A, et al. Labeling of enveloped virus via metabolic incorporation of azido sugars[J]. Bioconjugate Chemistry, 2015, 26(9): 1868-1872. |
| 86 | OUM Y H, DESAI T M, MARIN M, et al. Click labeling of unnatural sugars metabolically incorporated into viral envelope glycoproteins enables visualization of single particle fusion[J]. Journal of Virological Methods, 2016, 233: 62-71. |
| 87 | FERNANDEZ M V, FREED E O. Expand and click: a new method for labeling HIV-1 envelope glycoproteins [J]. Cell Chemical, Biology, 2017, 24 (5): 548-550. |
| 88 | JAO C Y, ROTH M, WELTI R, et al. Metabolic labeling and direct imaging of choline phospholipids in vivo [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(36): 15332-15337. |
| 89 | ANCAJAS C F, RICKS T J, BEST M D. Metabolic labeling of glycerophospholipids via clickable analogs derivatized at the lipid headgroup[J]. Chemistry and Physics of Lipids, 2020, 232: 104971. |
| 90 | BUMPUS T W, BASKIN J M. Greasing the wheels of lipid biology with chemical tools[J]. Trends in Biochemical Sciences, 2018, 43(12): 970-983. |
| 91 | HUANG L L, LU G H, HAO J, et al. Enveloped virus labeling via both intrinsic biosynthesis and metabolic incorporation of phospholipids in host cells[J]. Analytical Chemistry, 2013, 85(10): 5263-5270. |
| 92 | ZHAO X, SHEN Y, ADOGLA E A, et al. Surface labeling of enveloped virus with polymeric imidazole ligand-capped quantum dots via the metabolic incorporation of phospholipids into host cells[J]. Journal of Materials Chemistry B, 2016, 4(14): 2421-2427. |
| 93 | HAGEMEIJER M C, VONK A M, MONASTYRSKA I, et al. Visualizing coronavirus RNA synthesis in time by using click chemistry[J]. Journal of Virology, 2012, 86(10): 5808-5816. |
| 94 | KALVERAM B, LIHORADOVA O, INDRAN S V, et al. Using click chemistry to measure the effect of viral infection on host-cell RNA synthesis[J]. Journal of Visualized Experiments, 2013(78): 50809. |
| 95 | WANG I H, SUOMALAINEN M, ANDRIASYAN V, et al. Tracking viral genomes in host cells at single-molecule resolution[J]. Cell Host & Microbe, 2013, 14(4): 468-480. |
| 96 | SENKEVICH T G, KATSAFANAS G C, WEISBERG A, et al. Identification of vaccinia virus replisome and transcriptome proteins by isolation of proteins on nascent DNA coupled with mass spectrometry[J]. Journal of Virology, 2017, 91(19): e01015-e01017. |
| 97 | GIERLICH J, BURLEY G A, GRAMLICH P M E, et al. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA[J]. Organic Letters, 2006, 8(17): 3639-3642. |
| 98 | SALIC A, MITCHISON T J. A chemical method for fast and sensitive detection of DNA synthesis in vivo [J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(7): 2415-2420. |
| 99 | HUANG L L, LIU K J, ZHANG Q M, et al. Integrating two efficient and specific bioorthogonal ligation reactions with natural metabolic incorporation in one cell for virus dual labeling[J]. Analytical Chemistry, 2017, 89(21): 11620-11627. |
| 100 | LU Z P, XIAO Z L, YANG Z, et al. Hepatitis B virus X protein promotes human hepatoma cell growth via upregulation of transcription factor AP2α and sphingosine kinase 1[J]. Acta Pharmacologica Sinica, 2015, 36(10): 1228-1236. |
| 101 | RUBINO F A, OUM Y H, RAJARAM L, et al. Chemoselective modification of viral surfaces via bioorthogonal click chemistry[J]. Journal of Visualized Experiments: JoVE, 2012(66): e4246. |
| 102 | WANG I H, BURCKHARDT C J, YAKIMOVICH A, et al. Imaging, tracking and computational analyses of virus entry and egress with the cytoskeleton[J]. Viruses, 2018, 10(4): 166. |
| 103 | LIU Xiaohui, OUYANG Ting, OUYANG Hongsheng, et al. Single particle labeling of RNA virus in live cells [J]. Virus Research, 2017, 237: 14-21. |
| 104 | HART C, CHASE L G, HAJIVANDI M, et al. Metabolic labeling and click chemistry detection of glycoprotein markers of mesenchymal stem cell differentiation[M]// Mesenchymal stem cell assays and applications, Springer, 2011: 459-484. |
| 105 | LAUGHLIN S T, AGARD N J, BASKIN J M, et al. Metabolic labeling of glycans with azido sugars for visualization and glycoproteomics[J]. Methods in Enzymology, 2006, 415: 230-250. |
| 106 | ZHANG Xiu, ZHANG Yan. Applications of azide-based bioorthogonal click chemistry in glycobiology[J]. Molecules, 2013, 18(6): 7145-7159. |
| 107 | TEO S H C, SERWA R A, O'HARE P. Spatial and temporal resolution of global protein synthesis during HSV infection using bioorthogonal precursors and click chemistry[J]. PLoS Pathogens, 2016, 12(10): e1005927. |
| 108 | ALANDIJANY T, ROBERTS A P E, CONN K L, et al. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection[J]. PLoS Pathogens, 2018, 14(1): e1006769. |
| 109 | NAGAO M, FUJIWARA Y, MATSUBARA T, et al. Design of glycopolymers carrying sialyl oligosaccharides for controlling the interaction with the influenza virus[J]. Biomacromolecules, 2017, 18(12): 4385-4392. |
| 110 | GAO M, NETTLES R E, BELEMA M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect[J]. Nature, 2010, 465(7294): 96-100. |
| 111 | JONES L H, BEAL D, SELBY M D, et al. In-cell click labelling of small molecules to determine subcellular localisation[J]. Journal of Chemical Biology, 2011, 4(2): 49-53. |
| 112 | WANG C, ZHU W D, WANG B Z. Dual-linker gold nanoparticles as adjuvanting carriers for multivalent display of recombinant influenza hemagglutinin trimers and flagellin improve the immunological responses in vivo and in vitro [J]. International Journal of Nanomedicine, 2017, 12: 4747-4762. |
| 113 | PATEL K G, SWARTZ J R. Surface functionalization of virus-like particles by direct conjugation using azide-alkyne click chemistry[J]. Bioconjugate Chemistry, 2011, 22(3): 376-387. |
| 114 | VENTER P A, DIRKSEN A, THOMAS D, et al. Multivalent display of proteins on viral nanoparticles using molecular recognition and chemical ligation strategies[J]. Biomacromolecules, 2011, 12(6): 2293-2301. |
| 115 | THRANE S, JANITZEK C M, AGERBÆK M Ø, et al. A novel virus-like particle based vaccine platform displaying the placental malaria antigen VAR2CSA[J]. PLoS One, 2015, 10(11): e0143071. |
| 116 | KATO T, KAWAGUCHI A, NAGATA K, et al. Development of tetraphenylethylene-based fluorescent oligosaccharide probes for detection of influenza virus[J]. Biochemical and Biophysical Research Communications, 2010, 394(1): 200-204. |
| 117 | PFEILSTICKER J A, UMEDA A, FARROW B, et al. A cocktail of thermally stable, chemically synthesized capture agents for the efficient detection of anti-gp41 antibodies from human sera[J]. PLoS One, 2013, 8(10): e76224. |
| 118 | MATSUBARA T, UJIE M, YAMAMOTO T, et al. Highly sensitive detection of influenza virus by boron-doped diamond electrode terminated with sialic acid-mimic peptide[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(32): 8981-8984. |
| 119 | YUE G Y, YE H Z, HUANG X J, et al. Quantification of DNA through a fluorescence biosensor based on click chemistry[J]. The Analyst, 2014, 139(22): 5669-5673. |
| 120 | THOMSON D A C, COOPER M A. A paramagnetic-reporter two-particle system for amplification-free detection of DNA in serum[J]. Biosensors and Bioelectronics, 2013, 50: 499-501. |
| 121 | THOMSON D A C, TEE E H L, TRAN N T D, et al. Oligonucleotide and polymer functionalized nanoparticles for amplification-free detection of DNA[J]. Biomacromolecules, 2012, 13(6): 1981-1989. |
| 122 | SUN X X, CUI Z Q. Virus-like particles as theranostic platforms[J]. Advanced Therapeutics, 2020, 3(5): 1900194. |
| 123 | SUN Y P, LÜ X Q, DING P T, et al. Exploring the functions of polymers in adenovirus-mediated gene delivery: evading immune response and redirecting tropism[J]. Acta Biomaterialia, 2019, 97: 93-104. |
| 124 | HONG J, YUN C O. Overcoming the limitations of locally administered oncolytic virotherapy[J]. BMC Biomedical Engineering, 2019, 1: 17. |
| 125 | CHU Y J, OUM Y H, CARRICO I S. Surface modification via strain-promoted click reaction facilitates targeted lentiviral transduction[J]. Virology, 2016, 487: 95-103. |
| 126 | KELEMEN R E, MUKHERJEE R, CAO X F, et al. A precise chemical strategy to alter the receptor specificity of the adeno-associated virus[J]. Angewandte Chemie International Edition, 2016, 55(36): 10645-10649. |
| 127 | HUANG L L, LI X, LIU K J, et al. Engineering oncolytic vaccinia virus with functional peptides through mild and universal strategy[J]. Analytical and Bioanalytical Chemistry, 2019, 411(4): 925-933. |
| 128 | MANDHARE A, BANERJEE P, BHUTKAR S, et al. "Click chemistry" for diagnosis: a patent review on exploitation of its emerging trends[J]. Expert Opinion on Therapeutic Patents, 2014, 24(12): 1287-1310. |
| 129 | BRUCKMAN M A, KAUR G, LEE L A, et al. Surface modification of tobacco mosaic virus withclick chemistry[J]. ChemBioChem, 2008, 9(4): 519-523. |
| [1] | ZHANG Shouqi, WANG Tao, KONG Yao, ZOU Jiasheng, LIU Yuanning, XU Zhengren. Chemoenzymatic synthesis of natural products: evolution of synthetic methodology and strategy [J]. Synthetic Biology Journal, 2024, 5(5): 913-940. |
| [2] | CHENG Zhongyu, LI Fuzhuo. Recent advances in chemoenzymatic synthesis of natural products via site- selective P450 oxidation [J]. Synthetic Biology Journal, 2024, 5(5): 960-980. |
| [3] | GUO Xiya, CHEN Ji, DONG Mingxin. New strategies for engineering influenza viruses and their applications [J]. Synthetic Biology Journal, 2024, 5(2): 267-280. |
| [4] | Botao JI, Zhigang QIAN, Xiaoxia XIA. Application of cell-free synthesis strategy in biomaterial research [J]. Synthetic Biology Journal, 2022, 3(4): 658-675. |
| [5] | Wenjing ZHANG, Ming LI, Wei ZHOU, Xian-en ZHANG, Feng LI. Self-assembly, biosynthesis, functionalization and applications of virus-based nanomaterials [J]. Synthetic Biology Journal, 2020, 1(3): 298-318. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||