Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (5): 966-984.DOI: 10.12211/2096-8280.2022-012
• Invited Review • Previous Articles Next Articles
Applications of regulatory engineering in photosynthetic cyanobacteria
DONG Zhengxin1,3,4,5, SUN Tao1,2,4, CHEN Lei1,3,4,5, ZHANG Weiwen1,2,3,4,5
- 1.Laboratory of Synthetic Microbiology,School of Chemical Engineering & Technology,Tianjin University,Tianjin 300072,China
2.Center for Biosafety Research and Strategy,Tianjin University,Tianjin 300072,China
3.Key Laboratory of Systems Bioengineering,Ministry of Education of China,Tianjin 300072,China
4.Frontier Science Center for Synthetic Biology,Ministry of Education of China,Tianjin 300072,China
5.Collaborative Innovation Center of Chemical Science and Engineering,Tianjin 300072,China
-
Received:2022-02-11Revised:2022-04-01Online:2022-11-16Published:2022-10-31 -
Contact:SUN Tao, ZHANG Weiwen
调控工程在光合蓝细菌中的应用
董正鑫1,3,4,5, 孙韬1,2,4, 陈磊1,3,4,5, 张卫文1,2,3,4,5
- 1.天津大学化工学院合成微生物学实验室,天津 300072
2.天津大学生物安全战略研究中心,天津 300072
3.教育部系统生物工程重点实验室,天津 300072
4.教育部合成生物学前沿科学中心,天津 300072
5.化学化工协同创新中心,天津 300072
-
通讯作者:孙韬,张卫文 -
作者简介:董正鑫 (1997—),男,博士研究生。主要研究方向为微生物合成生物学。 E-mail:dongxin7122@foxmail.com孙韬 (1990—),博士,讲师,硕士生导师。从事微生物合成生物学及生物安全相关研究。 E-mail:tsun@tju.edu.cn张卫文 (1967—),男,博士,教授,博士生导师,生物安全战略研究中心主任。长期从事光合微生物的合成生物学,以及生物安全相关研究,牵头中美合成生物学生物安全“二轨”战略对话。 E-mail:wwzhang8@tju.edu.cn -
基金资助:国家重点研发计划(2018YFA0903000)
CLC Number:
Cite this article
DONG Zhengxin, SUN Tao, CHEN Lei, ZHANG Weiwen. Applications of regulatory engineering in photosynthetic cyanobacteria[J]. Synthetic Biology Journal, 2022, 3(5): 966-984.
董正鑫, 孙韬, 陈磊, 张卫文. 调控工程在光合蓝细菌中的应用[J]. 合成生物学, 2022, 3(5): 966-984.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-012
| 菌株 | 调控系统单元 | 所属调控系统 | 功能 |
|---|---|---|---|
| PCC 6803 | Hik31-Rre34 | TCS | 维持Cu2+稳态[ |
| PCC 6803 | Sll0649 | TCS | 维持Cu2+、Cd2+、Fe2+、Mn2+、Zn2+稳态[ |
| Nostocflagelliforme CCNUN1 | OrrA | TCS | 调控MAA合成,抵抗UV-B和干旱[ |
| PCC 6803 | Slr1037、Sll0039 | TCS | 丁醇耐受[ |
| PCC 6803 | Hik33、Hik34、Hik16、Hik41 | TCS | NaCl耐受[ |
| PCC 6803 | Hik2-Rre1 Hik34-Rre1 | TCS | NaCl耐受[ |
| PCC 6803 | Hik36-Hik43-Rre6 | TCS | NaCl耐受,生物膜生成[ |
| PCC 7942 | Synpcc7942_1125、Synpcc7942_1404 | TCS | NaCl耐受[ |
| PCC 7102 | OrrA | TCS | NaCl耐受、干旱[ |
| PCC 6803 | Rre37 | TCS | 氮缺乏、三羧酸循环、丙酮酸代谢、琥珀酸合成[ |
| PCC 7102 | SigB2 | σ因子 | NaCl耐受[ |
| PCC 6803 | SigE | σ因子 | 琥珀酸合成[ |
| PCC 6803 | SigB | σ因子 | 盐耐受、热耐受、丁醇耐受、氧化耐受[ |
| PCC 6803 | SigD | σ因子 | 氧化耐受[ |
| PCC 6803 | SigE | σ因子 | 糖原降解、光系统蛋白丰度、琥珀酸合成、聚羟基丁酸酯 合成[ |
| PCC 7120 | SigE | σ因子 | 氮固定[ |
| PCC 6803 | CoaR | sRNA | 丁醇耐受[ |
| PCC 6803 | IsrR | sRNA | 铁离子缺乏、氧化耐受[ |
| PCC 6803 | IsaR1 | sRNA | 铁离子缺乏、盐耐受[ |
| PCC 6803 | PsbA2R、PsbA3R | sRNA | 光适应[ |
| PCC 6803 | PsrR1 | sRNA | 光适应[ |
| PCC 6803 | RblR | sRNA | 光适应、碳固定[ |
| PCC 6803 | Yfr1 | sRNA | 碳固定、氧化耐受、盐耐受、活性氧耐受[ |
| PCC 6803 | Nc117 | sRNA | 醇类耐受[ |
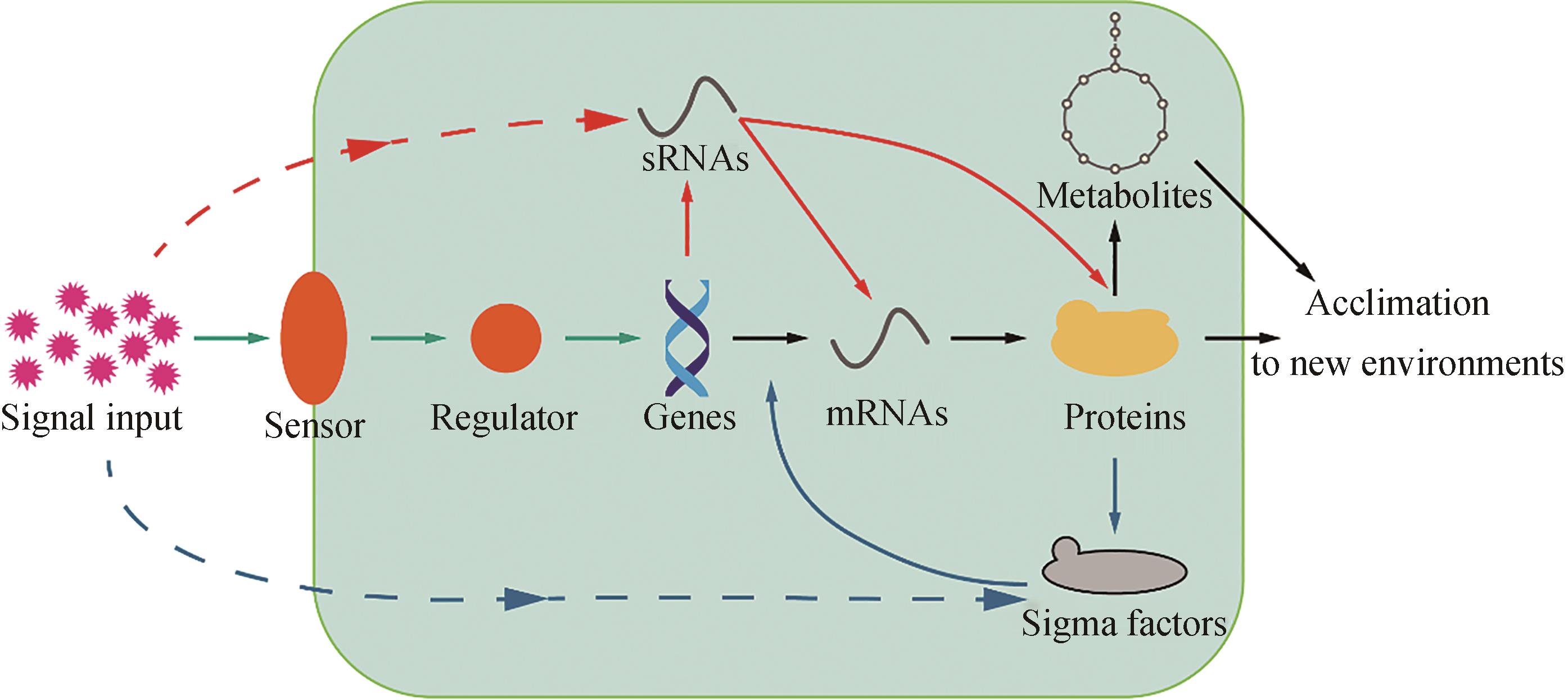
Tab. 1 Functions of regulatory system units in cyanobacteria
| 菌株 | 调控系统单元 | 所属调控系统 | 功能 |
|---|---|---|---|
| PCC 6803 | Hik31-Rre34 | TCS | 维持Cu2+稳态[ |
| PCC 6803 | Sll0649 | TCS | 维持Cu2+、Cd2+、Fe2+、Mn2+、Zn2+稳态[ |
| Nostocflagelliforme CCNUN1 | OrrA | TCS | 调控MAA合成,抵抗UV-B和干旱[ |
| PCC 6803 | Slr1037、Sll0039 | TCS | 丁醇耐受[ |
| PCC 6803 | Hik33、Hik34、Hik16、Hik41 | TCS | NaCl耐受[ |
| PCC 6803 | Hik2-Rre1 Hik34-Rre1 | TCS | NaCl耐受[ |
| PCC 6803 | Hik36-Hik43-Rre6 | TCS | NaCl耐受,生物膜生成[ |
| PCC 7942 | Synpcc7942_1125、Synpcc7942_1404 | TCS | NaCl耐受[ |
| PCC 7102 | OrrA | TCS | NaCl耐受、干旱[ |
| PCC 6803 | Rre37 | TCS | 氮缺乏、三羧酸循环、丙酮酸代谢、琥珀酸合成[ |
| PCC 7102 | SigB2 | σ因子 | NaCl耐受[ |
| PCC 6803 | SigE | σ因子 | 琥珀酸合成[ |
| PCC 6803 | SigB | σ因子 | 盐耐受、热耐受、丁醇耐受、氧化耐受[ |
| PCC 6803 | SigD | σ因子 | 氧化耐受[ |
| PCC 6803 | SigE | σ因子 | 糖原降解、光系统蛋白丰度、琥珀酸合成、聚羟基丁酸酯 合成[ |
| PCC 7120 | SigE | σ因子 | 氮固定[ |
| PCC 6803 | CoaR | sRNA | 丁醇耐受[ |
| PCC 6803 | IsrR | sRNA | 铁离子缺乏、氧化耐受[ |
| PCC 6803 | IsaR1 | sRNA | 铁离子缺乏、盐耐受[ |
| PCC 6803 | PsbA2R、PsbA3R | sRNA | 光适应[ |
| PCC 6803 | PsrR1 | sRNA | 光适应[ |
| PCC 6803 | RblR | sRNA | 光适应、碳固定[ |
| PCC 6803 | Yfr1 | sRNA | 碳固定、氧化耐受、盐耐受、活性氧耐受[ |
| PCC 6803 | Nc117 | sRNA | 醇类耐受[ |
| 65 | BILLIS K, BILLINI M, TRIPP H J, et al. Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation[J]. PLoS One, 2014, 9(10): e109738. |
| 66 | KOPF M, HESS W R. Regulatory RNAs in photosynthetic cyanobacteria[J]. FEMS Microbiology Reviews, 2015, 39(3): 301-315. |
| 67 | SUN T, CHEN L, ZHANG W W. Quantitative proteomics reveals potential crosstalk between a small RNA CoaR and a two-component regulator Slr1037 in Synechocystis sp. PCC6803[J]. Journal of Proteome Research, 2017, 16(8): 2954-2963. |
| 68 | WŁODARCZYK A, SELÃO T T, NORLING B, et al. Newly discovered Synechococcus sp. PCC 11901 is a robust cyanobacterial strain for high biomass production[J]. Communications Biology, 2020, 3: 215. |
| 69 | JAISWAL D, SENGUPTA A, SOHONI S, et al. Genome features and biochemical characteristics of a robust, fast growing and naturally transformable cyanobacterium Synechococcus elongatus PCC 11801 isolated from India[J]. Scientific Reports, 2018, 8: 16632. |
| 70 | KANNO M, CARROLL A L, ATSUMI S. Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria[J]. Nature Communications, 2017, 8: 14724. |
| 71 | KE T, LIU J, ZHAO S, et al. Using global transcription machinery engineering (GTME) and site-saturation mutagenesis technique to improve ethanol yield of Saccharomyces cerevisiae [J]. Applied Biochemistry and Microbiology, 2020, 56(5): 563-568. |
| 72 | YAMADA R, WAKITA K, OGINO H. Global metabolic engineering of glycolytic pathway via multicopy integration in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2017, 6(4): 659-666. |
| 73 | YAMADA R, WAKITA K, MITSUI R, et al. Enhanced d-lactic acid production by recombinant Saccharomyces cerevisiae following optimization of the global metabolic pathway[J]. Biotechnology and Bioengineering, 2017, 114(9): 2075-2084. |
| 74 | GINER-LAMIA J, LÓPEZ-MAURY L, REYES J C, et al. The CopRS two-component system is responsible for resistance to copper in the cyanobacterium Synechocystis sp. PCC 6803[J]. Plant Physiology, 2012, 159(4): 1806-1818. |
| 75 | GINER-LAMIA J, LÓPEZ-MAURY L, FLORENCIO F J. CopM is a novel copper-binding protein involved in copper resistance in Synechocystis sp. PCC 6803[J]. MicrobiologyOpen, 2015, 4(1): 167-185. |
| 76 | CHEN L, ZHU Y, SONG Z D, et al. An orphan response regulator Sll0649 involved in cadmium tolerance and metal homeostasis in photosynthetic Synechocystis sp. PCC 6803[J]. Journal of Proteomics, 2014, 103: 87-102. |
| 77 | GERALDES V, PINTO E. Mycosporine-like amino acids (MAAs): biology, chemistry and identification features[J]. Pharmaceuticals, 2021, 14(1): 63. |
| 1 | BRUCKNER B, HUBACEK K, SHAN Y L, et al. Impacts of poverty alleviation on national and global carbon emissions[J]. Nature Sustainability, 2022, 5(4): 311-320. |
| 2 | SHIH C F, ZHANG T, LI J H, et al. Powering the future with liquid sunshine[J]. Joule, 2018, 2(10): 1925-1949. |
| 3 | 李树斌, 孙韬, 陈磊, 等. 聚球藻UTEX 2973中光碳驱动的高密度燃料合成[J]. 生物工程学报, 2020, 36(10): 2126-2140. |
| LI S B, SUN T, CHEN L, et al. Light and carbon dioxide-driven synthesis of high-density fuel in Synechococcus elongatus UTEX 2973[J]. Chinese Journal of Biotechnology, 2020, 36(10): 2126-2140. | |
| 4 | KHAN F, SHAHID A, ZHU H, et al. Prospects of algae-based green synthesis of nanoparticles for environmental applications[J]. Chemosphere, 2022, 293: 133571. |
| 5 | KATO Y, INABE K, HIDESE R, et al. Metabolomics-based engineering for biofuel and bio-based chemical production in microalgae and cyanobacteria: a review[J]. Bioresource Technology, 2022, 344: 126196. |
| 6 | JAISWAL D, SAHASRABUDDHE D, WANGIKAR P P. Cyanobacteria as cell factories: the roles of host and pathway engineering and translational research[J]. Current Opinion in Biotechnology, 2022, 73: 314-322. |
| 7 | LIU X F, MIAO R, LINDBERG P, et al. Modular engineering for efficient photosynthetic biosynthesis of 1-butanol from CO2 in cyanobacteria[J]. Energy & Environmental Science, 2019, 12(9): 2765-2777. |
| 8 | KNOOT C J, PAKRASI H B. Diverse hydrocarbon biosynthetic enzymes can substitute for olefin synthase in the cyanobacterium Synechococcus sp. PCC 7002[J]. Scientific Reports, 2019, 9(1): 1360. |
| 9 | ZHANG L, CHEN L, DIAO J J, et al. Construction and analysis of an artificial consortium based on the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 to produce the platform chemical 3-hydroxypropionic acid from CO2 [J]. Biotechnology for Biofuels, 2020, 13: 82. |
| 10 | KOCH M, BRUCKMOSER J, SCHOLL J, et al. Maximizing PHB content in Synechocystis sp. PCC 6803: a new metabolic engineering strategy based on the regulator PirC[J]. Microbial Cell Factories, 2020, 19(1): 231. |
| 11 | VAN ALPHEN P, ABEDINI NAJAFABADI H, BRANCO DOS SANTOS F, et al. Increasing the photoautotrophic growth rate of Synechocystis sp. PCC 6803 by identifying the limitations of its cultivation[J]. Biotechnology Journal, 2018, 13(8): e1700764. |
| 12 | YU J J, LIBERTON M, CLIFTEN P F, et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2 [J]. Scientific Reports, 2015, 5: 8132. |
| 13 | GIBSON B, WILSON D J, FEIL E, et al. The distribution of bacterial doubling times in the wild[J]. Proceedings Biological Sciences, 2018, 285(1880): 20180789. |
| 14 | SALARI R, SALARI R. Investigation of the best Saccharomyces cerevisiae growth condition[J]. Electronic Physician, 2017, 9(1): 3592-3597. |
| 15 | XIE Y R, CHEN L, SUN T, et al. Deciphering and engineering high-light tolerant cyanobacteria for efficient photosynthetic cell factories[J]. Chinese Journal of Chemical Engineering, 2021, 30: 82-91. |
| 16 | CUI J Y, SUN T, LI S B, et al. Improved salt tolerance and metabolomics analysis of Synechococcus elongatus UTEX 2973 by overexpressing mrp antiporters[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 500. |
| 17 | LIU S Q, QURESHI N, HUGHES S R. Progress and perspectives on improving butanol tolerance[J]. World Journal of Microbiology & Biotechnology, 2017, 33(3): 51. |
| 18 | XIE Y R, CHEN L, SUN T, et al. Transporters related to stress responses and their potential application in Synechocystis sp. PCC 6803[M]// ZHANGWW, SONGXY. Synthetic biology of cyanobacteria[M]// Advances in experimental medicine and biology (Volume 1080). Singapore: Springer Singapore. 2018: 27-53. |
| 19 | CHEN T T, WANG X N, ZHUANG L, et al. Development and optimization of a microbial co-culture system for heterologous indigo biosynthesis[J]. Microbial Cell Factories, 2021, 20(1): 154. |
| 20 | JIA H Y, SUN X Y, SUN H, et al. Intelligent microbial heat-regulating engine (IMHeRE) for improved thermo-robustness and efficiency of bioconversion[J]. ACS Synthetic Biology, 2016, 5(4): 312-320. |
| 21 | BAO S H, JIANG H, ZHU L Y, et al. A dynamic and multilocus metabolic regulation strategy using quorum-sensing-controlled bacterial small RNA[J]. Cell Reports, 2021, 36(3): 109413. |
| 22 | CUI J Y, SUN T, CHEN L, et al. Engineering salt tolerance of photosynthetic cyanobacteria for seawater utilization[J]. Biotechnology Advances, 2020, 43: 107578. |
| 23 | LI X Q, SHEN C R, LIAO J C. Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942[J]. Photosynthesis Research, 2014, 120(3): 301-310. |
| 78 | SHANG J L, ZHANG Z C, YIN X Y, et al. UV-B induced biosynthesis of a novel sunscreen compound in solar radiation and desiccation tolerant cyanobacteria[J]. Environmental Microbiology, 2018, 20(1): 200-213. |
| 79 | RAJ S, KUNIYIL A M, SREENIKETHANAM A, et al. Microalgae as a source of mycosporine-like amino acids (MAAs): advances and future prospects[J]. International Journal of Environmental Research and Public Health, 2021, 18(23): 12402. |
| 80 | NIU X F, ZHU Y, PEI G S, et al. Elucidating butanol tolerance mediated by a response regulator Sll0039 in Synechocystis sp. PCC 6803 using a metabolomic approach[J]. Applied Microbiology and Biotechnology, 2015, 99(4): 1845-1857. |
| 81 | MARIN K, SUZUKI I, YAMAGUCHI K, et al. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(15): 9061-9066. |
| 82 | VIDAL R, LÓPEZ-MAURY L, GUERRERO M G, et al. Characterization of an alcohol dehydrogenase from the cyanobacterium Synechocystis sp. strain PCC 6803 that responds to environmental stress conditions via the Hik34-Rre1 two-component system[J]. Journal of Bacteriology, 2009, 191(13): 4383-4391. |
| 83 | LIANG C W, ZHANG X W, CHI X Y, et al. Serine/threonine protein kinase SpkG is a candidate for high salt resistance in the unicellular cyanobacterium Synechocystis sp. PCC 6803[J]. PLoS One, 2011, 6(5): e18718. |
| 84 | VIDAL R. Identification of the correct form of the mis-annotated response regulator Rre1 from the cyanobacterium Synechocystis sp. PCC 6803[J]. FEMS Microbiology Letters, 2015, 362(7): fnv030. |
| 85 | IBRAHIM I M, PUTHIYAVEETIL S, ALLEN J F. A two-component regulatory system in transcriptional control of photosystem stoichiometry: redox-dependent and sodium ion-dependent phosphoryl transfer from cyanobacterial histidine kinase Hik2 to response regulators Rre1 and RppA[J]. Frontiers in Plant Science, 2016, 7: 137. |
| 86 | KERA K, YOSHIZAWA Y, SHIGEHARA T, et al. Hik36-Hik43 and Rre6 act as a two-component regulatory system to control cell aggregation in Synechocystis sp. PCC6803[J]. Scientific Reports, 2020, 10: 19405. |
| 87 | QIAO C C, ZHANG M Y, LUO Q, et al. Identification of two two-component signal transduction mutants with enhanced sucrose biosynthesis in Synechococcus elongatus PCC 7942[J]. Journal of Basic Microbiology, 2019, 59(5): 465-476. |
| 88 | EHIRA S, KIMURA S, MIYAZAKI S, et al. Sucrose synthesis in the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 is controlled by the two-component response regulator OrrA[J]. Applied and Environmental Microbiology, 2014, 80(18): 5672-5679. |
| 89 | KIMURA S, SATO M, FAN X Y, et al. The two-component response regulator OrrA confers dehydration tolerance by regulating anaKa expression in the cyanobacterium Anabaena sp. strain PCC 7120[EB/OL]. bioRxiv, 2021. DOI: 10.1101/2021.08.03.454875 . |
| 24 | WU X X, LI J W, XING S F, et al. Establishment of a resource recycling strategy by optimizing isobutanol production in engineered cyanobacteria using high salinity stress[J]. Biotechnology for Biofuels, 2021, 14(1): 174. |
| 25 | KLÄHN S, MIKKAT S, RIEDIGER M, et al. Integrative analysis of the salt stress response in cyanobacteria[J]. Biology Direct, 2021, 16(1): 26. |
| 26 | GAYSINA L A, SARAF A, SINGH P. Chapter 1-Cyanobacteria in Diverse Habitats [M]// MISHRA A K, TIWARI D N, RAI A N. Cyanobacteria. Academic Press, 2019: 1-28. |
| 27 | RACHEDI R, FOGLINO M, LATIFI A. Stress signaling in cyanobacteria: a mechanistic overview[J]. Life, 2020, 10(12): E312. |
| 28 | MIRONOV K S, SINETOVA M A, SHUMSKAYA M, et al. Universal molecular triggers of stress responses in cyanobacterium Synechocystis [J]. Life, 2019, 9(3): 67. |
| 29 | HU J L, WANG Q. Regulatory sRNAs in cyanobacteria[J]. Frontiers in Microbiology, 2018, 9: 2399. |
| 30 | KIM I M, SZURMANT H. A bacterial Goldilocks mechanism[J]. eLife, 2020, 9: e54244. |
| 31 | MIRA-RODADO V. New insights into multistep-phosphorelay (MSP)/two-component system (TCS) regulation: are plants and bacteria that different? [J]. Plants, 2019, 8(12): 590. |
| 32 | KRAKOWIAK J, ZHENG X, PATEL N, et al. Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response[J]. eLife, 2018, 7: e31668. |
| 33 | LI G T, MORIGEN, YAO Y. TorR/TorS Two-component system resists extreme acid environment by regulating the key response factor RpoS in Escherichia coli [J]. Gene, 2022, 821: 146295. |
| 34 | PENG F, CHEN J, LIU X X, et al. The PhoPR two-component system responds to oxygen deficiency and regulates the pathways for energy supply in Corynebacterium glutamicum [J]. World Journal of Microbiology & Biotechnology, 2021, 37(9): 160. |
| 35 | KANEKO T, SATO S, KOTANI H, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803 (Ⅱ): Sequence determination of the entire genome and assignment of potential protein-coding regions[J]. DNA Research, 1996, 3(3): 185-209. |
| 90 | IIJIMA H, WATANABE A, TAKANOBU J, et al. rre37 Overexpression alters gene expression related to the tricarboxylic acid cycle and pyruvate metabolism in Synechocystis sp. PCC 6803[J]. The Scientific World Journal, 2014, 2014: 921976. |
| 91 | TAKEYA M, IIJIMA H, SUKIGARA H, et al. Cluster-level relationships of genes involved in carbon metabolism in Synechocystis sp. PCC 6803: development of a novel succinate-producing strain[J]. Plant and Cell Physiology, 2017, 59(1): 72-81. |
| 92 | TYYSTJÄRVI T, HUOKKO T, RANTAMÄKI S, et al. Impact of different group 2 sigma factors on light use efficiency and high salt stress in the cyanobacterium Synechocystis sp. PCC 6803[J]. PLoS One, 2013, 8(4): e63020. |
| 93 | KACZMARZYK D, ANFELT J, SÄRNEGRIM A, et al. Overexpression of sigma factor SigB improves temperature and butanol tolerance of Synechocystis sp. PCC6803[J]. Journal of Biotechnology, 2014, 182/183: 54-60. |
| 94 | TOKUMARU Y, UEBAYASHI K, TOYOSHIMA M, et al. Comparative targeted proteomics of the central metabolism and photosystems in SigE mutant strains of Synechocystis sp. PCC 6803[J]. Molecules, 2018, 23(5): 1051. |
| 95 | OSANAI T, NUMATA K, OIKAWA A, et al. Increased bioplastic production with an RNA polymerase sigma factor SigE during nitrogen starvation in Synechocystis sp. PCC 6803[J]. DNA Research, 2013, 20(6): 525-535. |
| 96 | GRUBER T M, BRYANT D A. Characterization of the alternative σ-factors SigD and SigE in Synechococcus sp. strain PCC 7002. SigE is implicated in transcription of post-exponential-phase-specific genes[J]. Archives of Microbiology, 1998, 169(3): 211-219. |
| 97 | MELLA-HERRERA R A, NEUNUEBEL M R, KUMAR K, et al. The sigE gene is required for normal expression of heterocyst-specific genes in Anabaena sp. strain PCC 7120[J]. Journal of Bacteriology, 2011, 193(8): 1823-1832. |
| 98 | LI S B, SUN T, CHEN L, et al. Designing and constructing artificial small RNAs for gene regulation and carbon flux redirection in photosynthetic cyanobacteria[J]. Methods in Molecular Biology, 2021, 2290: 229-252. |
| 99 | SINGH A K, SHERMAN L A. Reflections on the function of IsiA, a cyanobacterial stress-inducible, Chl-binding protein[J]. Photosynthesis Research, 2007, 93(1/2/3): 17-25. |
| 100 | DÜHRING U, AXMANN I M, HESS W R, et al. An internal antisense RNA regulates expression of the photosynthesis gene isiA [J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(18): 7054-7058. |
| 101 | GEORG J, KOSTOVA G, VUORIJOKI L, et al. Acclimation of oxygenic photosynthesis to iron starvation is controlled by the sRNA IsaR1[J]. Current Biology, 2017, 27(10): 1425-1436.e7. |
| 36 | SUGITA C, OGATA K, SHIKATA M, et al. Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: gene content and organization[J]. Photosynthesis Research, 2007, 93(1/2/3): 55-67. |
| 37 | GAO X Y, SUN T, WU L N, et al. Co-overexpression of response regulator genes slr1037 and sll0039 improves tolerance of Synechocystis sp. PCC 6803 to 1-butanol[J]. Bioresource Technology, 2017, 245: 1476-1483. |
| 38 | CHEN L, WU L N, WANG J X, et al. Butanol tolerance regulated by a two-component response regulator Slr1037 in photosynthetic Synechocystis sp. PCC 6803[J]. Biotechnology for Biofuels, 2014, 7: 89. |
| 39 | REN Q, SHI M L, CHEN L, et al. Integrated proteomic and metabolomic characterization of a novel two-component response regulator Slr1909 involved in acid tolerance in Synechocystis sp. PCC 6803[J]. Journal of Proteomics, 2014, 109: 76-89. |
| 40 | SHI M L, CHEN L, ZHANG W W. Regulatory diversity and functional analysis of two-component systems in cyanobacterium Synechocystis sp. PCC 6803 by GC-MS based metabolomics[J]. Frontiers in Microbiology, 2020, 11: 403. |
| 41 | PEI G S, NIU X F, ZHOU Y Q, et al. Crosstalk of two-component signal transduction systems in regulating central carbohydrate and energy metabolism during autotrophic and photomixotrophic growth of Synechocystis sp. PCC 6803[J]. Integrative Biology, 2017, 9(5): 485-496. |
| 42 | SHEN C H. Gene expression: transcription of the genetic code[M]// SHEN C H. Diagnostic molecular biology. Amsterdam: Elsevier, 2019: 59-86. |
| 43 | RIAZ-BRADLEY A. Transcription in cyanobacteria: a distinctive machinery and putative mechanisms[J]. Biochemical Society Transactions, 2019, 47(2): 679-689. |
| 44 | SRIVASTAVA A, SUMMERS M L, SOBOTKA R. Cyanobacterial sigma factors: current and future applications for biotechnological advances[J]. Biotechnology Advances, 2020, 40: 107517. |
| 45 | SUN D, LIU C, ZHU J R, et al. Connecting metabolic pathways: sigma factors in streptomyces spp[J]. Frontiers in Microbiology, 2017, 8: 2546. |
| 46 | PAGET M S. Bacterial sigma factors and anti-sigma factors: structure, function and distribution[J]. Biomolecules, 2015, 5(3): 1245-1265. |
| 47 | MAZUMDER A, KAPANIDIS A N. Recent advances in understanding σ70-dependent transcription initiation mechanisms[J]. Journal of Molecular Biology, 2019, 431(20): 3947-3959. |
| 102 | RÜBSAM H, KIRSCH F, REIMANN V, et al. The iron-stress activated RNA 1 (IsaR1) coordinates osmotic acclimation and iron starvation responses in the cyanobacterium Synechocystis sp. PCC 6803[J]. Environmental Microbiology, 2018, 20(8): 2757-2768. |
| 103 | SAKURAI I, STAZIC D, EISENHUT M, et al. Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803[J]. Plant Physiology, 2012, 160(2): 1000-1010. |
| 104 | GEORG J, DIENST D, SCHÜRGERS N, et al. The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria[J]. The Plant Cell, 2014, 26(9): 3661-3679. |
| 105 | NAKAMURA T, NAITO K, YOKOTA N, et al. A cyanobacterial non-coding RNA, Yfr1, is required for growth under multiple stress conditions[J]. Plant and Cell Physiology, 2007, 48(9): 1309-1318. |
| 106 | LIN W R, TAN S I, HSIANG C C, et al. Challenges and opportunity of recent genome editing and multi-omics in cyanobacteria and microalgae for biorefinery[J]. Bioresource Technology, 2019, 291: 121932. |
| 107 | KU J T, LAN E I. Design principles for engineering metabolic pathways in cyanobacteria[M]// Cyanobacteria biotechnology. Springer, 2021: 211-235. |
| 108 | LIN P C, PAKRASI H B. Engineering cyanobacteria for production of terpenoids[J]. Planta, 2019, 249(1): 145-154. |
| 109 | XU K, QIN L, BAI W X, et al. Multilevel defense system (MDS) relieves multiple stresses for economically boosting ethanol production of industrial Saccharomyces cerevisiae [J]. ACS Energy Letters, 2020, 5(2): 572-582. |
| 110 | KATAYAMA N, IIJIMA H, OSANAI T. Production of bioplastic compounds by genetically manipulated and metabolic engineered cyanobacteria// ZHANG W W, SONG X Y. Synthetic biology of cyanobacteria[M]// Advances in experimental medicine and biology (Volume 1080). Singapore: Springer Singapore. 2018: 155-169. |
| 111 | SUN T, LI S B, SONG X Y, et al. Toolboxes for cyanobacteria: recent advances and future direction[J]. Biotechnology Advances, 2018, 36(4): 1293-1307. |
| 112 | CHEN L, WU L N, ZHU Y, et al. An orphan two-component response regulator Slr1588 involves salt tolerance by directly regulating synthesis of compatible solutes in photosynthetic Synechocystis sp. PCC 6803[J]. Molecular BioSystems, 2014, 10(7): 1765-1774. |
| 113 | LIU P, ZHANG B, YAO Z H, et al. Multiplex design of the metabolic network for production of L-homoserine in Escherichia coli [J]. Applied and Environmental Microbiology, 2020, 86(20): e01477-e01420. |
| 48 | DAVIS M C, KESTHELY C A, FRANKLIN E A, et al. The essential activities of the bacterial sigma factor[J]. Canadian Journal of Microbiology, 2017, 63(2): 89-99. |
| 49 | KOSKINEN S, HAKKILA K, GUNNELIUS L, et al. In vivo recruitment analysis and a mutant strain without any group 2 σ factor reveal roles of different σ factors in cyanobacteria[J]. Molecular Microbiology, 2016, 99(1): 43-54. |
| 50 | TURUNEN O, KOSKINEN S, KURKELA J, et al. Roles of close homologues SigB and SigD in heat and high light acclimation of the cyanobacterium Synechocystis sp. PCC 6803[J]. Life, 2022, 12(2): 162. |
| 51 | HAKKILA K, VALEV D, ANTAL T, et al. Group 2 σ factors are central regulators of oxidative stress acclimation in cyanobacteria[J]. Plant and Cell Physiology, 2018, 60(2): 436-447. |
| 52 | FLORES C, SANTOS M, PEREIRA S B, et al. The alternative sigma factor SigF is a key player in the control of secretion mechanisms in Synechocystis sp. PCC 6803[J]. Environmental Microbiology, 2019, 21(1): 343-359. |
| 53 | KIRSCH F, KLÄHN S, HAGEMANN M. Salt-regulated accumulation of the compatible solutes sucrose and glucosylglycerol in cyanobacteria and its biotechnological potential[J]. Frontiers in Microbiology, 2019, 10: 2139. |
| 54 | FLEMING K E, O'SHEA E K. An RpaA-dependent sigma factor cascade sets the timing of circadian transcriptional rhythms in Synechococcus elongatus [J]. Cell Reports, 2018, 25(11): 2937-2945.e3. |
| 55 | HASEGAWA H, TSURUMAKI T, KOBAYASHI I, et al. Identification and analysis of a principal sigma factor interacting protein SinA, essential for growth at high temperatures in a cyanobacterium Synechococcus elongatus PCC 7942[J]. The Journal of General and Applied Microbiology, 2020, 66(2): 66-72. |
| 56 | ZAMPETAKI A, ALBRECHT A, STEINHOFEL K. Long non-coding RNA structure and function: is there a link? [J]. Frontiers in Physiology, 2018, 9: 1201. |
| 57 | O'BRIEN J, HAYDER H, ZAYED Y, et al. Overview of microRNA biogenesis, mechanisms of actions, and circulation[J]. Frontiers in Endocrinology, 2018, 9: 402. |
| 58 | HOLMQVIST E, VOGEL J. RNA-binding proteins in bacteria[J]. Nature Reviews Microbiology, 2018, 16(10): 601-615. |
| 59 | GEORG J, VOSS B, SCHOLZ I, et al. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation[J]. Molecular Systems Biology, 2009, 5: 305. |
| 114 | LAPINAITE A, KNOTT G J, PALUMBO C M, et al. DNA capture by a CRISPR-Cas9-guided adenine base editor[J]. Science, 2020, 369(6503): 566-571. |
| 115 | MOK B Y, DE MORAES M H, ZENG J, et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing[J]. Nature, 2020, 583(7817): 631-637. |
| 116 | WATANABE S. Cyanobacterial multi-copy chromosomes and their replication[J]. Bioscience, Biotechnology, and Biochemistry, 2020, 84(7): 1309-1321. |
| 117 | OHBAYASHI R, WATANABE S, KANESAKI Y, et al. DNA replication depends on photosynthetic electron transport in cyanobacteria[J]. FEMS Microbiology Letters, 2013, 344(2): 138-144. |
| 118 | OHBAYASHI R, YAMAMOTO J Y, WATANABE S, et al. Variety of DNA replication activity among cyanobacteria correlates with distinct respiration activity in the dark[J]. Plant and Cell Physiology, 2016, 58(2): 279-286. |
| 119 | OHBAYASHI R, WATANABE S, EHIRA S, et al. Diversification of DnaA dependency for DNA replication in cyanobacterial evolution[J]. The ISME Journal, 2016, 10(5): 1113-1121. |
| 120 | KOBAYASHI S, ATSUMI S, IKEBUKURO K, et al. Light-induced production of isobutanol and 3-methyl-1-butanol by metabolically engineered cyanobacteria[J]. Microbial Cell Factories, 2022, 21(1): 7. |
| 121 | KUKIL K, LINDBERG P. Expression of phenylalanine ammonia lyases in Synechocystis sp. PCC 6803 and subsequent improvements of sustainable production of phenylpropanoids[J]. Microbial Cell Factories, 2022, 21(1): 8. |
| 122 | 许可, 王靖楠, 李春. 智能抗逆微生物细胞工厂与绿色生物制造[J]. 合成生物学, 2020, 1(4): 427-439. |
| XU K, WANG J N, LI C. Intelligent microbial cell factory with tolerance for green biological manufacturing[J]. Synthetic Biology Journal, 2020, 1(4): 427-439. | |
| 123 | DU Q, LIU Y L, SONG Y Y, et al. Creation of a low-alcohol-production yeast by a mutated SPT15 transcription regulator triggers transcriptional and metabolic changes during wine fermentation[J]. Frontiers in Microbiology, 2020, 11: 597828. |
| 124 | ALPER H, MOXLEY J, NEVOIGT E, et al. Engineering yeast transcription machinery for improved ethanol tolerance and production[J]. Science, 2006, 314(5805): 1565-1568. |
| 125 | YANG K K, WU Z, ARNOLD F H. Machine-learning-guided directed evolution for protein engineering[J]. Nature Methods, 2019, 16(8): 687-694. |
| 60 | SUN T, PEI G S, WANG J X, et al. A novel small RNA CoaR regulates coenzyme A biosynthesis and tolerance of Synechocystis sp. PCC6803 to 1-butanol possibly via promoter-directed transcriptional silencing[J]. Biotechnology for Biofuels, 2017, 10: 42. |
| 61 | HU J L, LI T P, XU W, et al. Small antisense RNA RblR positively regulates RuBisCo in Synechocystis sp. PCC 6803[J]. Frontiers in Microbiology, 2017, 8: 231. |
| 62 | BI Y Q, PEI G S, SUN T, et al. Regulation mechanism mediated by trans-encoded sRNA Nc117 in short chain alcohols tolerance in Synechocystis sp. PCC 6803[J]. Frontiers in Microbiology, 2018, 9: 863. |
| 63 | TAN X M, HOU S W, SONG K, et al. The primary transcriptome of the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973[J]. Biotechnology for Biofuels, 2018, 11: 218. |
| 64 | PEI G S, SUN T, CHEN S, et al. Systematic and functional identification of small non-coding RNAs associated with exogenous biofuel stress in cyanobacterium Synechocystis sp. PCC 6803[J]. Biotechnology for Biofuels, 2017, 10: 57. |
| 126 | LI C Y, ZHANG R H, WANG J, et al. Protein engineering for improving and diversifying natural product biosynthesis[J]. Trends in Biotechnology, 2020, 38(7): 729-744. |
| 127 | YU KING HING N, ARYAL U K, MORGAN J A. Probing light-dependent regulation of the Calvin cycle using a multi-omics approach[J]. Frontiers in Plant Science, 2021, 12: 733122. |
| 128 | SANTOS-MERINO M, TORRADO A, DAVIS G A, et al. Improved photosynthetic capacity and photosystem I oxidation via heterologous metabolism engineering in cyanobacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(11): e2021523118. |
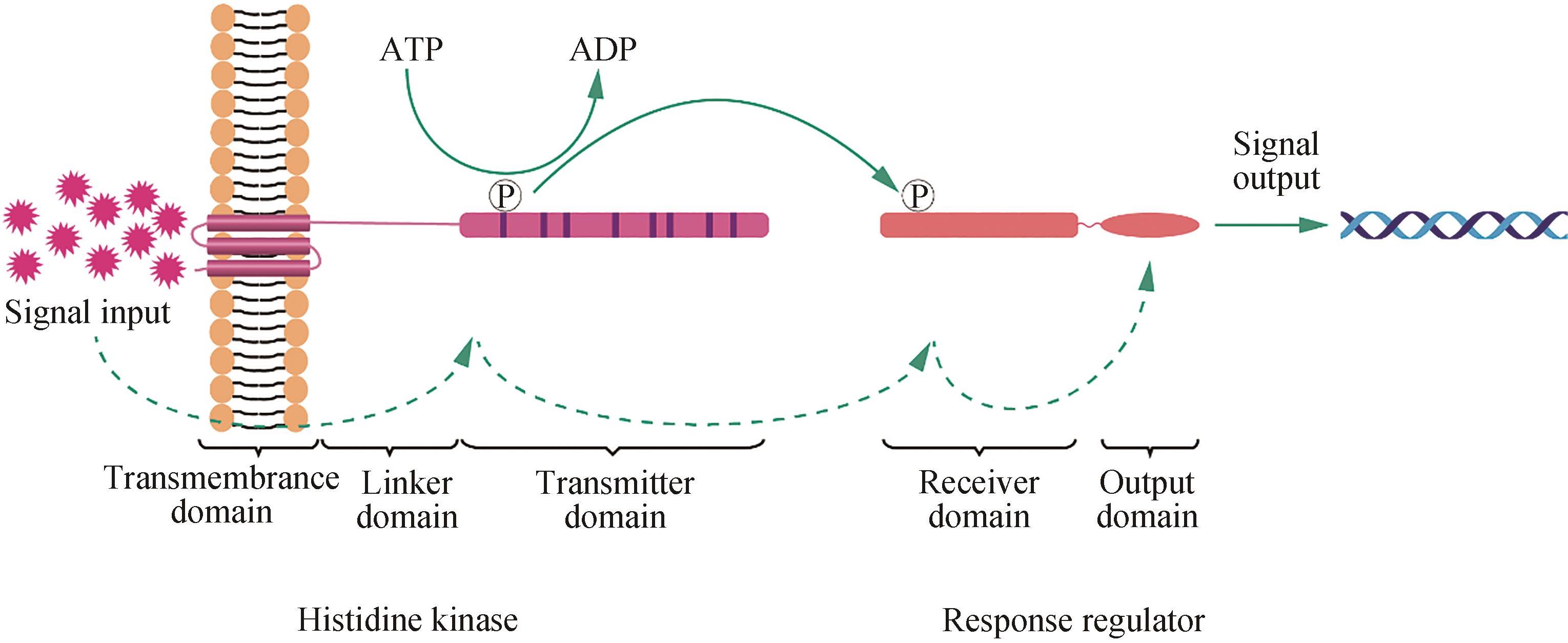
| [1] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [2] | Zhen ZHU, Jing TIAN, Jing JIANG, Wangyin WANG, Xupeng CAO. Progress in microalgae chloroplast organelle factory development [J]. Synthetic Biology Journal, 2022, 3(6): 1218-1234. |
| [3] | YANG Jianzhao, ZHU Xinguang. Plant synthetic biology for carbon peak and carbon neutrality [J]. Synthetic Biology Journal, 2022, 3(5): 847-869. |
| [4] | Menglin SHI, Lin ZHOU, Qing WANG, Lei ZHAO. Advances in the study on the modification of carbon dioxide metabolic pathways in plants [J]. Synthetic Biology Journal, 2022, 3(5): 985-1005. |
| [5] | TAO Fei, SUN Tao, WANG Yu, WEI Ting, NI Jun, XU Ping. Challenges and opportunities in the research of Synechococcus chassis under the context of carbon peak and neutrality [J]. Synthetic Biology Journal, 2022, 3(5): 932-952. |
| [6] | ZHAO Quanyu. Research progress in carbon neutrality oriented adaptive laboratory evolution of microalgae [J]. Synthetic Biology Journal, 2022, 3(5): 901-914. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||